
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,429,432 – 18,429,533 |
| Length | 101 |
| Max. P | 0.711605 |

| Location | 18,429,432 – 18,429,533 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Shannon entropy | 0.47976 |
| G+C content | 0.49911 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -13.47 |
| Energy contribution | -13.19 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18429432 101 - 23011544 --GGGCGAGGGAAACACAUUCUGUCAGUCAUUUGGCAGCUCUGGCCAACCAUUGUCACAACAUUUGUGCAA--UCUCUCUCGCUUUCCUGACAUUUUCCCCCGUU- --.((((.((((((.......((((((....(((((.......)))))..((((.((((.....)))))))--).............)))))).)))))).))))- ( -26.40, z-score = -1.27, R) >droSim1.chr2L 18117035 101 - 22036055 --AGGCGAGGGAAACACAUUCUGUCAGUCAUUUGGCAGCUCUGGCCAACCAUUGUCACAACAUUUGUGCAA--UCUCUCUCGCUUUCCCGACAUUUUCCCCCGUU- --((((((((((....((..(((((((....)))))))...)).......((((.((((.....)))))))--).))))))))))....................- ( -26.40, z-score = -1.71, R) >droSec1.super_7 2052460 101 - 3727775 --AGGCGAGGGAAACACAUUCUGUCAGUCAUUUGGCAGCUCUGGCCAACCAUUGUCACAACAUUUGUGCAA--UCUCUCUCGCUUUCCCGACAUUUUCCCCCGUU- --((((((((((....((..(((((((....)))))))...)).......((((.((((.....)))))))--).))))))))))....................- ( -26.40, z-score = -1.71, R) >droYak2.chr2R 4945469 101 - 21139217 --AGGCGAGGGAAACACAUUCUGUCAGUCAUUUGGCAGCUCUGGCCAACCAUUGUCACAACAUUUGUGCCA--UCUCUCUCGAUUUUCCGACAUUUUCCCCCGUU- --.((((.((((((......(((((((....)))))))...(((((((....(((....))).))).))))--......(((......)))...)))))).))))- ( -25.20, z-score = -1.31, R) >droEre2.scaffold_4845 16744343 103 + 22589142 --AGGCGAGGGAAACACAUUCUGUCAGUCAUUUGGCAGCUCUGGCCAACCAUUGUCACAACAUUUGCGCCAUCUCUCUCUCGCUUUUCCGACAUUUUCUCCCGCU- --((((((((((........(((((((....)))))))...(((((((....(((....))).))).))))....))))))))))....................- ( -27.70, z-score = -2.26, R) >droAna3.scaffold_12916 2337296 106 - 16180835 AGAUGGGCCAGAAGCAUAUUCUGUCAGUCAGUUGGCGUCCCUGGCAAAGCAUUGUCACAACAUUUCCAUAUUUUCGUUCUCUUUCGGCAAUGUUUUUCCCUCUCUU (((.(((((((..((.....(((.....)))...))....)))))(((((((((((.............................))))))))))).)).)))... ( -24.65, z-score = -1.60, R) >dp4.chr4_group3 7332048 87 + 11692001 ----------GACACACAUUCUGUCAGUCAUUUGGCAACCCUGGCCAACCAUUGUCACAACAUUUCUGCAGAGUC-CACGCCCUGCUCUCCCCCUGUC-------- ----------((((.......((.((((...(((((.......)))))..)))).))..........((((.((.-...)).))))........))))-------- ( -16.20, z-score = -0.46, R) >droPer1.super_1 4440246 88 + 10282868 ---------AGACACACAUUCUGUCAGUCAUUUGGCAACCCUGGCCAACCAUUGUCACAACAUUUCUGCAGAGUC-CACGCACUGCUCCCCCCCUGUC-------- ---------.((((.......((.((((...(((((.......)))))..)))).))..........((((.((.-...)).))))........))))-------- ( -16.30, z-score = -0.15, R) >consensus __AGGCGAGGGAAACACAUUCUGUCAGUCAUUUGGCAGCUCUGGCCAACCAUUGUCACAACAUUUGUGCAA__UCUCUCUCGCUUUUCCGACAUUUUCCCCCGUU_ ....(((.((((((.......((((((....))))))....((((........)))).....................................)))))).))).. (-13.47 = -13.19 + -0.28)

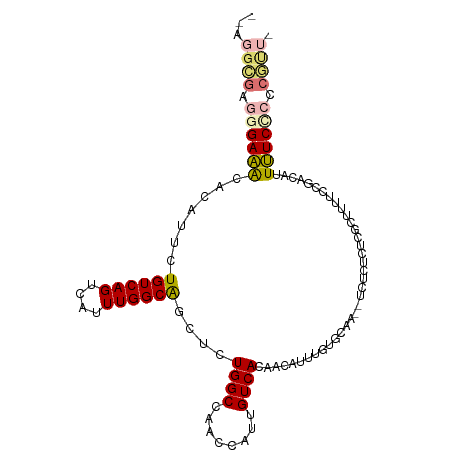
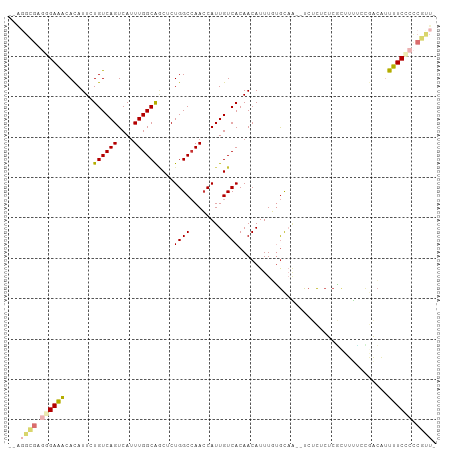
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:03 2011