| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,414,268 – 18,414,337 |
| Length | 69 |
| Max. P | 0.845624 |

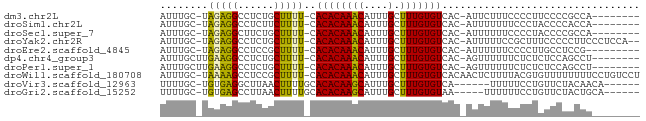
| Location | 18,414,268 – 18,414,337 |
|---|---|
| Length | 69 |
| Sequences | 10 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 74.41 |
| Shannon entropy | 0.51354 |
| G+C content | 0.41547 |
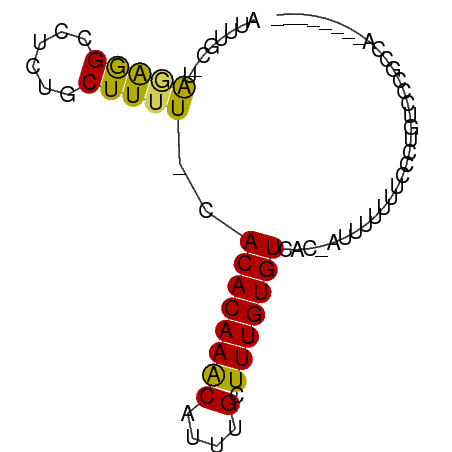
| Mean single sequence MFE | -10.67 |
| Consensus MFE | -6.64 |
| Energy contribution | -5.92 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.845624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18414268 69 - 23011544 AUUUGC-UAGAGGCCUCUGCUUUU-CACACAAACAUUUGCUUUGUGUCAC-AUUCUUUCCCCUUCCCCGCCA-------- ....((-.((((((....))))))-.((((((((....).)))))))...-.................))..-------- ( -9.00, z-score = -0.83, R) >droSim1.chr2L 18101493 69 - 22036055 AUUUGC-UAGAGGCCUCUUCUUUU-CACACAAACAUUUGCUUUGUGUCAC-AUUUUUUUCCCUACCCCACCA-------- ......-...(((...........-.((((((((....).)))))))...-.........))).........-------- ( -6.85, z-score = -0.86, R) >droSec1.super_7 2037346 69 - 3727775 AUUUGC-UAGAGGCUUCUGCUUUU-CACACAAACAUUUGCUUUGUGUCAC-AUUUUUUCCCCUACCCCGCCA-------- ....((-.((((((....))))))-.((((((((....).)))))))...-.................))..-------- ( -10.20, z-score = -1.42, R) >droYak2.chr2R 4929118 75 - 21139217 AUUUGC-UAGAGGCCUCUGCUUUU-CACACAAACAUUUGCUUUGUGUCAC-AUUUUUCCGCUUUCCCCCUUCCCUCCA-- ....((-.((((((....))))))-.((((((((....).)))))))...-........)).................-- ( -9.40, z-score = -1.24, R) >droEre2.scaffold_4845 16729282 68 + 22589142 AUUUGC-UAGAGGCCUCCGCUUUU-CACACAAACAUUUGCUUUGUGUCAC-AUUUUUUCCCCUUGCCUCCG--------- ......-..(((((..........-.((((((((....).)))))))...-.............)))))..--------- ( -10.43, z-score = -1.71, R) >dp4.chr4_group3 7310420 70 + 11692001 AUUUGCUUGAAGGCCUCUGCUUUU-CACACAAACAUUUGCUUUGUGUCAC-AGUUUUUUCUCUCUCCAGCCU-------- ....(((.((..(.....(((...-.((((((((....).)))))))...-))).......)..)).)))..-------- ( -10.20, z-score = -0.20, R) >droPer1.super_1 4418615 70 + 10282868 AUUUGCUUGAAGGCCUCUGCUUUU-CACACAAACAUUUGCUUUGUGUCAC-AGUUUUUUCUCUCUCCAGCCU-------- ....(((.((..(.....(((...-.((((((((....).)))))))...-))).......)..)).)))..-------- ( -10.20, z-score = -0.20, R) >droWil1.scaffold_180708 11534620 78 - 12563649 AUUUGC-UAAAAGCCUCCGCUUUU-CACACAAACAUUUGCUUUGUGUCACAACUCUUUUACGUGUUUUUUUUCCUGUCCU ......-.((((((....))))))-.((((((((....).)))))))................................. ( -8.20, z-score = -0.92, R) >droVir3.scaffold_12963 18368046 67 - 20206255 UUUUGC-UGUGAGGCUUAACUUUUGCACACAAGCAUUUGCUUUGUGUCA------UUUUUCCUGUUCUACAACA------ ...((.-((((..((........((.((((((((....)).))))))))------........))..)))).))------ ( -14.49, z-score = -1.26, R) >droGri2.scaffold_15252 6359414 68 - 17193109 UUUUGC-UGUGAGCCUUAACUUUUGCACACAAGCAUUUGCUUUGUGUAA-----UUUUUUCCUGUUCUACUGCA------ ...(((-.((((((........(((((((.((((....)))))))))))-----.........)))).)).)))------ ( -17.73, z-score = -3.07, R) >consensus AUUUGC_UAGAGGCCUCUGCUUUU_CACACAAACAUUUGCUUUGUGUCAC_AUUUUUUUCCCUGUCCCGCCA________ ........(((((......)))))..((((((((....).)))))))................................. ( -6.64 = -5.92 + -0.72)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:02 2011