| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,401,804 – 18,401,898 |
| Length | 94 |
| Max. P | 0.829687 |

| Location | 18,401,804 – 18,401,898 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.90 |
| Shannon entropy | 0.58373 |
| G+C content | 0.37641 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -9.23 |
| Energy contribution | -9.70 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
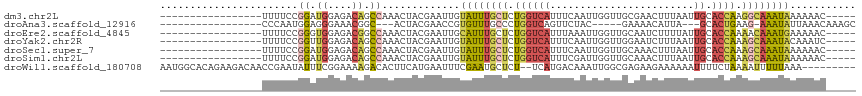
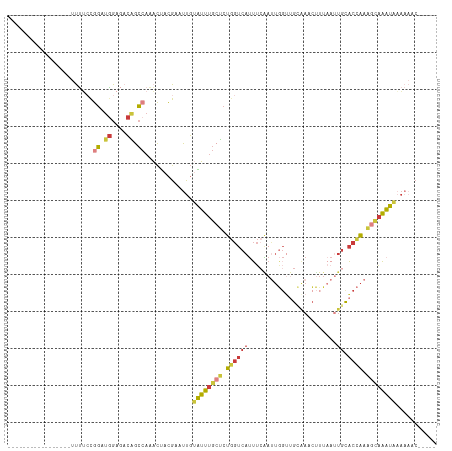
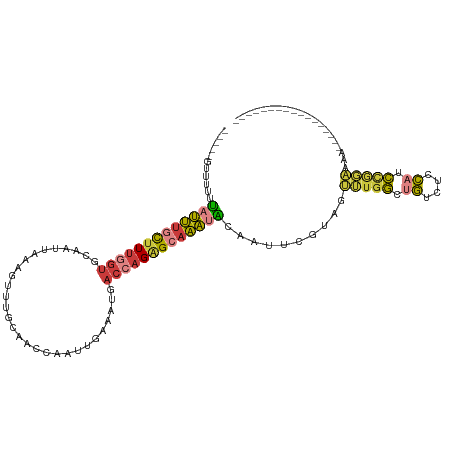
>dm3.chr2L 18401804 94 + 23011544 -----------------UUUUCCGGAUGGAGACAGCCAAACUACGAAUUGUAUUUGCUCUGGUCAUUUCAAUUGGUUGCGAACUUUAAUUGCACCAAGGCAAAUAAAAAAC----- -----------------......((.((....)).)).............(((((((..((((.....(((((((.........))))))).))))..)))))))......----- ( -21.20, z-score = -1.36, R) >droAna3.scaffold_12916 2313166 87 + 16180835 -----------------CCCAAUGGAGGGAAACGGC---ACUACGAACCGUGUUUGCCCUGGUCAGUUCUAC-----GAAAACAUUA---GCACUGAAG-AAAUAUUAAACAAAGC -----------------(((......)))....(((---(.((((...))))..))))....(((((.(((.-----........))---).)))))..-................ ( -15.30, z-score = 0.42, R) >droEre2.scaffold_4845 16721637 94 - 22589142 -----------------UUUUCCGGGUGGAGACGGCCAAACUACGAAUUGCAUUUGCUCUGGUCAUUUAAAUUGGUUGCAAUCUUUUAUUGCACCAAAACAAAUGAAAAAC----- -----------------....((((.((....)).))............((....))...))((((((...(((((.(((((.....))))))))))...)))))).....----- ( -22.60, z-score = -1.63, R) >droYak2.chr2R 4919296 94 + 21139217 -----------------UUUUCCGGUUGGAGACAGCCAAACUACGAAUUGUAUUUGCUCUGGUCAUUUCAAUUGGUUGGAAUCUUUAAUUGCACCAAAGCAAAUACAAAUC----- -----------------......(((((....))))).......((.(((((((((((.((((((((((((....))))))........)).)))).))))))))))).))----- ( -30.90, z-score = -4.75, R) >droSec1.super_7 2029654 94 + 3727775 -----------------UUUUCCGGAUGGAGACAGCCAAACUACGAAUUGUAUUUGCUCUGGUCAUUUCAAUUGGUUGCAAACUUUAAUUGCACCAAAGCAAAUAAAAAAC----- -----------------......((.((....)).)).............((((((((.(((.....))).(((((.((((.......)))))))))))))))))......----- ( -21.40, z-score = -1.89, R) >droSim1.chr2L 18093783 94 + 22036055 -----------------UUUUCCGGAUGGAGACAGCCAAACUACGAAUUGUAUUUGCUCUGGUCAUUUCGAUUGGUUGCAAACUUUAAUUGCACCAAAGCAAAUAAAAAAC----- -----------------......((.((....)).)).............((((((((.((((((.......))...((((.......)))))))).))))))))......----- ( -20.80, z-score = -1.54, R) >droWil1.scaffold_180708 11523116 105 + 12563649 AAUGGCACAGAAGACAACCGAAUAUUUCGGAAAAGACACUUCAUGAAUUUCGAAUGCUCU--UCAUGACAAAUUGGCGAGAAGAAAAAAUUUUCUAAAAUUUUUAAA--------- .......(((((((((..((((...(((((((.......))).)))).))))..)).)))--)).))..(((((....((((((.....))))))..))))).....--------- ( -15.90, z-score = -0.06, R) >consensus _________________UUUUCCGGAUGGAGACAGCCAAACUACGAAUUGUAUUUGCUCUGGUCAUUUCAAUUGGUUGCAAACUUUAAUUGCACCAAAGCAAAUAAAAAAC_____ .......................((.((....)).)).............((((((((.((((((........................)).)))).))))))))........... ( -9.23 = -9.70 + 0.48)
| Location | 18,401,804 – 18,401,898 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.90 |
| Shannon entropy | 0.58373 |
| G+C content | 0.37641 |
| Mean single sequence MFE | -20.89 |
| Consensus MFE | -9.98 |
| Energy contribution | -10.19 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
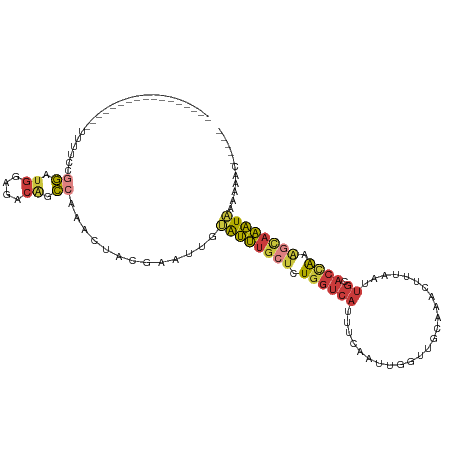
>dm3.chr2L 18401804 94 - 23011544 -----GUUUUUUAUUUGCCUUGGUGCAAUUAAAGUUCGCAACCAAUUGAAAUGACCAGAGCAAAUACAAUUCGUAGUUUGGCUGUCUCCAUCCGGAAAA----------------- -----(..((.(((((((.(((((.(((((.............))))).....))))).))))))).))..)....(((((.((....)).)))))...----------------- ( -16.02, z-score = 0.49, R) >droAna3.scaffold_12916 2313166 87 - 16180835 GCUUUGUUUAAUAUUU-CUUCAGUGC---UAAUGUUUUC-----GUAGAACUGACCAGGGCAAACACGGUUCGUAGU---GCCGUUUCCCUCCAUUGGG----------------- ((((((..........-..(((((.(---((.((....)-----)))).))))).))))))....(((((.......---)))))..(((......)))----------------- ( -18.42, z-score = -0.26, R) >droEre2.scaffold_4845 16721637 94 + 22589142 -----GUUUUUCAUUUGUUUUGGUGCAAUAAAAGAUUGCAACCAAUUUAAAUGACCAGAGCAAAUGCAAUUCGUAGUUUGGCCGUCUCCACCCGGAAAA----------------- -----.....(((((((..((((((((((.....))))).)))))..)))))))(((((.....(((.....))).)))))(((........)))....----------------- ( -23.00, z-score = -1.77, R) >droYak2.chr2R 4919296 94 - 21139217 -----GAUUUGUAUUUGCUUUGGUGCAAUUAAAGAUUCCAACCAAUUGAAAUGACCAGAGCAAAUACAAUUCGUAGUUUGGCUGUCUCCAACCGGAAAA----------------- -----((.((((((((((((((((.((.((...((((......)))).)).)))))))))))))))))).))....(((((.((....)).)))))...----------------- ( -28.80, z-score = -4.05, R) >droSec1.super_7 2029654 94 - 3727775 -----GUUUUUUAUUUGCUUUGGUGCAAUUAAAGUUUGCAACCAAUUGAAAUGACCAGAGCAAAUACAAUUCGUAGUUUGGCUGUCUCCAUCCGGAAAA----------------- -----(..((.(((((((((((((((((.......))))...((.......))))))))))))))).))..)....(((((.((....)).)))))...----------------- ( -22.80, z-score = -1.74, R) >droSim1.chr2L 18093783 94 - 22036055 -----GUUUUUUAUUUGCUUUGGUGCAAUUAAAGUUUGCAACCAAUCGAAAUGACCAGAGCAAAUACAAUUCGUAGUUUGGCUGUCUCCAUCCGGAAAA----------------- -----(..((.(((((((((((((((((.......))))...((.......))))))))))))))).))..)....(((((.((....)).)))))...----------------- ( -22.80, z-score = -1.83, R) >droWil1.scaffold_180708 11523116 105 - 12563649 ---------UUUAAAAAUUUUAGAAAAUUUUUUCUUCUCGCCAAUUUGUCAUGA--AGAGCAUUCGAAAUUCAUGAAGUGUCUUUUCCGAAAUAUUCGGUUGUCUUCUGUGCCAUU ---------...((((((((....))))))))..............((.(((((--(((.((..((((.(((..((((.....)))).)))...))))..))))))).))).)).. ( -14.40, z-score = 0.68, R) >consensus _____GUUUUUUAUUUGCUUUGGUGCAAUUAAAGUUUGCAACCAAUUGAAAUGACCAGAGCAAAUACAAUUCGUAGUUUGGCUGUCUCCAUCCGGAAAA_________________ ...........(((((((((((((.............................)))))))))))))..........(((((.((....)).))))).................... ( -9.98 = -10.19 + 0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:50:02 2011