
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,768,785 – 1,768,876 |
| Length | 91 |
| Max. P | 0.977198 |

| Location | 1,768,785 – 1,768,876 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 89.28 |
| Shannon entropy | 0.20012 |
| G+C content | 0.39049 |
| Mean single sequence MFE | -15.42 |
| Consensus MFE | -13.38 |
| Energy contribution | -13.62 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
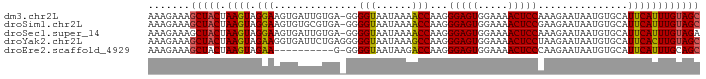
>dm3.chr2L 1768785 91 + 23011544 GCUACAAAUGAAUGCACAUUAUUCUUUGGAGUUUUCCACUCCCUUGGUUUUAUUACCCC-UCACAAUCACUUCCUACUUAGUAGCUUUCUUU (((((....(((((.....)))))...(((((.....)))))...(((......)))..-....................)))))....... ( -15.30, z-score = -2.86, R) >droSim1.chr2L 1755656 91 + 22036055 GCUACAAAUGAAUGCACAUUAUUCUUCGGAGUUUUCCACUCCCUUGGUUUUAUUACCCC-UCACGCACACUUCCUACUUAGUAGCUUUCUUU (((((....(((((.....)))))...(((((.....)))))...(((......)))..-....................)))))....... ( -15.60, z-score = -3.00, R) >droSec1.super_14 1703216 91 + 2068291 UCUACAAAUGAAUGCACAUUAUUCUUUGGAGUUUUCCACUCCCUUGGUUUUAUUACCCC-UCACAAUCACUUCCUACUUAGUAGCUUUCUUU .........(((.((((..........(((((.....)))))...(((......)))..-....................)).)).)))... ( -11.60, z-score = -1.39, R) >droYak2.chr2L 1742007 92 + 22324452 GCUACAAGUGAAUGCACAUUAUUCUUAGGAGUUUUCCACUCCCUUGGCUUUAUUACCCCCUCAGAAUCACCUUCUACUUAGUAGCUUUCUUU ((((((((((((((.....)))))...(((((.....)))))....................((((.....)))))))).)))))....... ( -16.40, z-score = -1.72, R) >droEre2.scaffold_4929 1813034 81 + 26641161 GCUGCAAAUGAAUGCACAUUAUUCUUGGGAGUUUUCCACUCCCUUGGUCUUAUUACCCC-C----------UUCUACUUAGUAGCUUUCUUU (((((....(((((.....)))))..((((((.....))))))..(((......)))..-.----------.........)))))....... ( -18.20, z-score = -2.80, R) >consensus GCUACAAAUGAAUGCACAUUAUUCUUUGGAGUUUUCCACUCCCUUGGUUUUAUUACCCC_UCACAAUCACUUCCUACUUAGUAGCUUUCUUU (((((....(((((.....)))))...(((((.....)))))...(((......))).......................)))))....... (-13.38 = -13.62 + 0.24)


| Location | 1,768,785 – 1,768,876 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 89.28 |
| Shannon entropy | 0.20012 |
| G+C content | 0.39049 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.941935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
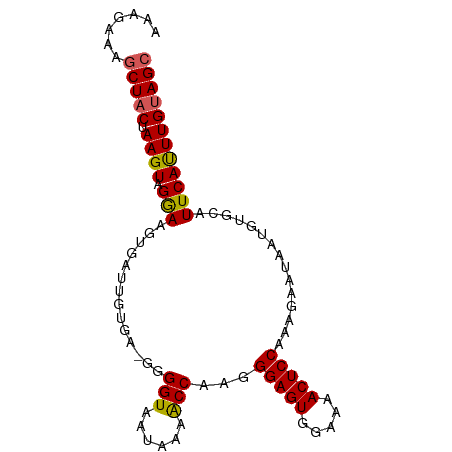
>dm3.chr2L 1768785 91 - 23011544 AAAGAAAGCUACUAAGUAGGAAGUGAUUGUGA-GGGGUAAUAAAACCAAGGGAGUGGAAAACUCCAAAGAAUAAUGUGCAUUCAUUUGUAGC .......(((((........((((((.(((..-..(((......)))...(((((.....)))))............))).))))))))))) ( -20.40, z-score = -2.55, R) >droSim1.chr2L 1755656 91 - 22036055 AAAGAAAGCUACUAAGUAGGAAGUGUGCGUGA-GGGGUAAUAAAACCAAGGGAGUGGAAAACUCCGAAGAAUAAUGUGCAUUCAUUUGUAGC .......(((((.((((.((..(((..(((..-..(((......)))...(((((.....)))))........)))..)))))))))))))) ( -23.30, z-score = -3.34, R) >droSec1.super_14 1703216 91 - 2068291 AAAGAAAGCUACUAAGUAGGAAGUGAUUGUGA-GGGGUAAUAAAACCAAGGGAGUGGAAAACUCCAAAGAAUAAUGUGCAUUCAUUUGUAGA ........((((...)))).((((((.(((..-..(((......)))...(((((.....)))))............))).))))))..... ( -18.40, z-score = -1.78, R) >droYak2.chr2L 1742007 92 - 22324452 AAAGAAAGCUACUAAGUAGAAGGUGAUUCUGAGGGGGUAAUAAAGCCAAGGGAGUGGAAAACUCCUAAGAAUAAUGUGCAUUCACUUGUAGC .......(((((.((((.(((.((.(((((.....(((......)))..((((((.....)))))).))))).....)).)))))))))))) ( -24.80, z-score = -2.70, R) >droEre2.scaffold_4929 1813034 81 - 26641161 AAAGAAAGCUACUAAGUAGAA----------G-GGGGUAAUAAGACCAAGGGAGUGGAAAACUCCCAAGAAUAAUGUGCAUUCAUUUGCAGC ........((((...))))..----------.-..(((......)))..((((((.....))))))..........((((......)))).. ( -18.40, z-score = -1.72, R) >consensus AAAGAAAGCUACUAAGUAGGAAGUGAUUGUGA_GGGGUAAUAAAACCAAGGGAGUGGAAAACUCCAAAGAAUAAUGUGCAUUCAUUUGUAGC .......(((((.((((.(((.((...........(((......)))...(((((.....)))))............)).)))))))))))) (-16.40 = -16.64 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:08 2011