
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,372,061 – 18,372,169 |
| Length | 108 |
| Max. P | 0.535060 |

| Location | 18,372,061 – 18,372,169 |
|---|---|
| Length | 108 |
| Sequences | 8 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.29 |
| Shannon entropy | 0.62279 |
| G+C content | 0.40607 |
| Mean single sequence MFE | -27.19 |
| Consensus MFE | -9.80 |
| Energy contribution | -10.75 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
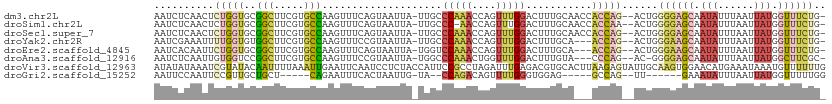
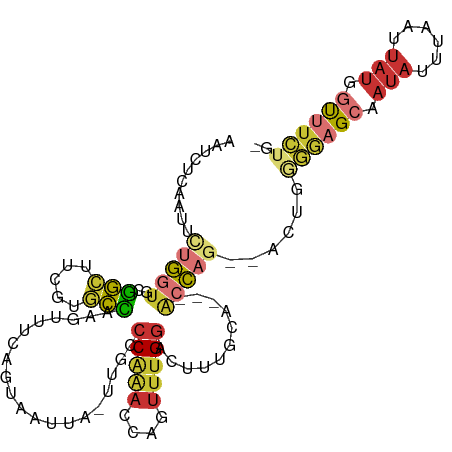
>dm3.chr2L 18372061 108 - 23011544 AAUCUCAACUCUGGUGCGGCUUCGUGCCAAGUUUCAGUAAUUA-UUGCCCAAACCAGUUUGGACUUUGCAACCACCAG--ACUGGGGAGCAAUAUUUAAUUAUGGUUUCUG- ......((((.(((..((....))..)))))))(((.(((.((-((((((...(((((((((............))))--))))))).)))))).)))....)))......- ( -32.70, z-score = -2.44, R) >droSim1.chr2L 18060246 107 - 22036055 AAUCUCAACUCUGGUGCGGCUUCGUGCCAAGUUUCAGUAAUUA-UUGCCC-AACCAGUUUGGACUUUGCAACCACCAA--ACUGGGGAGCAAUAUUUAAUUAUGGUUUCUG- ......((((.(((..((....))..)))))))(((.(((.((-((((((-..(((((((((............))))--))))))).)))))).)))....)))......- ( -33.00, z-score = -2.84, R) >droSec1.super_7 1998723 108 - 3727775 AAUCUCAACUCUGGUGCGGCUUCGUGCCAAGUUUCAGUAAUUA-UUGCCCAAACCAGUUUGGACUUUGCAACCACCAG--ACUGGGGAGCAAUAUUUAAUUAUGGUUUCUG- ......((((.(((..((....))..)))))))(((.(((.((-((((((...(((((((((............))))--))))))).)))))).)))....)))......- ( -32.70, z-score = -2.44, R) >droYak2.chr2R 4889222 105 - 21139217 AAUCGAAAUUUUGGUGUGGCUUCGUGCCAAGUUUCCGUAAUUA-UUGCCCAAACCAGUUUGGACUUUGCA---ACCAG--ACUGGGAAGCAAUAUUUAAUUAUGGUUUCUG- ....((((.(((((..((....))..)))))...(((((((((-((((.....(((((((((........---.))))--)))))...))))))....)))))))))))..- ( -32.90, z-score = -3.17, R) >droEre2.scaffold_4845 16692386 105 + 22589142 AAUCACAAUUCUGGUGCGGCUUCGUGCCAAGUUUCAGUAAUUA-UGGUCCAAACCAGUUUGGACUUUGCA---ACCAG--ACUGGGAAGCAAUAUUUAAUUAUGGUUUCUG- .........(((((((((((.....)))...............-.((((((((....))))))))..)).---)))))--)...((((((.(((......))).)))))).- ( -31.90, z-score = -2.78, R) >droAna3.scaffold_12916 2286140 104 - 16180835 AAUCUCAAUUGUGGUCCGGCUUCGUGCCAAGUUUCCGUAAUUA-UGGCCCAAACUGGUUUGGACUUUGUA---CCCAG--AC-GGGGAGCAAUAUUUAAUUAUGGCUUCGC- ..........((((...(((.....)))......(((((((((-.((.(((((....))))).))((((.---(((..--..-.))).))))....)))))))))..))))- ( -25.70, z-score = 0.19, R) >droVir3.scaffold_12963 18310837 112 - 20206255 AUAUAUAAAUCGUAUACAAUUUUAAAUUGAAUUCAAUCCUCUACCAUUCCGCCUAGAUUUGAGACGUGCACUUAAGAGUAUUGCAAGUGGAACAUGAAAUAAAUGUUUUUUG ..........(((...((((.....))))..(((((...((((..........)))).))))))))..(((((...........)))))((((((.......)))))).... ( -10.00, z-score = 2.05, R) >droGri2.scaffold_15252 6316872 91 - 17193109 AAUUCCAAUUCCGUUGCUGCU-----CAGAAUUUCACUAAUUG-UA--CCAGACAGUUUUGGGUGGAG-----GCCAG--UU------GAAAUAUUUAAUUAUGGUUUUUGG ....((((..((((..(..((-----(((((((((........-..--...)).)))))))))..)..-----)).((--((------((.....))))))..))...)))) ( -18.62, z-score = -0.11, R) >consensus AAUCUCAAUUCUGGUGCGGCUUCGUGCCAAGUUUCAGUAAUUA_UUGCCCAAACCAGUUUGGACUUUGCA___ACCAG__ACUGGGGAGCAAUAUUUAAUUAUGGUUUCUG_ ..........(((((..(((.....)))....................(((((....)))))...........)))))......((((((.(((......))).)))))).. ( -9.80 = -10.75 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:55 2011