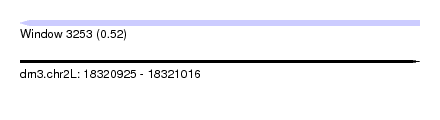
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,320,925 – 18,321,016 |
| Length | 91 |
| Max. P | 0.518995 |

| Location | 18,320,925 – 18,321,016 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 50.43 |
| Shannon entropy | 1.09679 |
| G+C content | 0.42413 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | 0.00 |
| Energy contribution | 0.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 0.00 |
| Mean z-score | -2.18 |
| Structure conservation index | -0.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.518995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
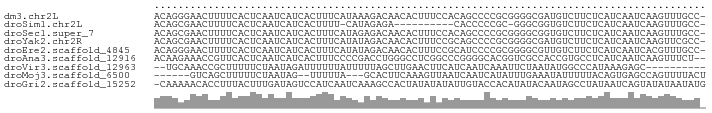
>dm3.chr2L 18320925 91 - 23011544 ACAGGGAACUUUUCACUCAAUCAUCACUUUCAUAAAGACAACACUUUCCACAGCCCCGCGGGGCGAUGUCUUCUCAUCAAUCAAGUUUGCC- ...(.((((((.........................................((((....))))((((......))))....)))))).).- ( -17.60, z-score = -1.61, R) >droSim1.chr2L 18008473 79 - 22036055 ACAGCGAACUUUUCACUCAAUCAUCACUUUU-CAUAGAGA----------CACCCCCGC-GGGCGGUGUCUUCUCAUCAAUCAAGUUUGCC- ...((((((((...............((...-...))(((----------((((((...-.)).)))))))...........)))))))).- ( -22.00, z-score = -4.18, R) >droSec1.super_7 1947472 91 - 3727775 ACAGCGAACUUUUCACUCAAUCAUCACUUUCAUAGAGACAACACUUUCCACAGCCCCGCGGGGCGGUGUCUUCUCAUCAAUCAAGUUUGCC- ...((((((((.......((........))....((((..(((((.......((((....)))))))))..)))).......)))))))).- ( -22.41, z-score = -3.27, R) >droYak2.chr2R 4838971 91 - 21139217 ACAGCGAACUUUUCACUCAAUCAUCACUUUCAUAUAGACAACACUUUCCGCAGCCCCGCGGGGCGAUGUCUUCUCAUCAAUCAAGUUCGCC- ...((((((((.........................................((((....))))((((......))))....)))))))).- ( -23.50, z-score = -4.51, R) >droEre2.scaffold_4845 16642334 91 + 22589142 ACAGGGAACUUUUCACUCAAUCAUCACUUUCAUAUAGACAACACUUUCCGCAUCCCCGCGGGGCGUUGUCUUCUCAUCAAUCACGUUUGCC- ...((((............................(((((((.((..((((......)))))).)))))))))))................- ( -17.18, z-score = -1.79, R) >droAna3.scaffold_12916 2239357 90 - 16180835 ACAAGAAACCGUUCACUCAAUCAUCACUUUCCCCGACCUGGGCCUCGGCCCGGGGCACGGUCGCCACCGUGCCUCAUCAAUCAAGUUUCU-- ...((((((..............................((((....))))((((((((((....)))))))))).........))))))-- ( -32.70, z-score = -4.20, R) >droVir3.scaffold_12963 18238541 80 - 20206255 --UGCAAACCGCUUUUUCUAAUAGAUUUUUUAUUUUUAGCUUGAACUUCAUCAAUCAAAUUCUAAUAUGGCCCAUAAAGAGC---------- --........(((((((((((.((((.....)))))))).((((......)))).....................)))))))---------- ( -5.30, z-score = 1.59, R) >droMoj3.scaffold_6500 26966747 81 - 32352404 ------GUCAGCUUUUUCUAAUAG--UUUUUA---GCACUUCAAAGUUAAUCAAUCAUAUUUGAAAUAUUUUUACAGUGAGCCAGUUUUACU ------....(((....((((...--...)))---)((((..(((((...((((......))))...)))))...))))))).......... ( -8.60, z-score = -0.08, R) >droGri2.scaffold_15252 6257922 91 - 17193109 -CAAAAACACCUUUACUUUGAUAGUCCAUCAAUCAAAGCCACUAUAUAUAUUGUACCACAUAUACAAUAGCCUAUAAUCAGUAUAUAAUAUG -..............(((((((.(.....).)))))))......(((((((((.........................)))))))))..... ( -10.01, z-score = -1.55, R) >consensus ACAGCGAACUUUUCACUCAAUCAUCACUUUCAUAUAGACAACACUUUCCACAGCCCCGCGGGGCGAUGUCUUCUCAUCAAUCAAGUUUGCC_ ............................................................................................ ( 0.00 = 0.00 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:49 2011