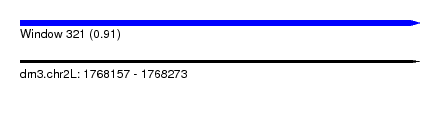
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,768,157 – 1,768,273 |
| Length | 116 |
| Max. P | 0.906409 |

| Location | 1,768,157 – 1,768,273 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Shannon entropy | 0.45766 |
| G+C content | 0.46357 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -10.76 |
| Energy contribution | -10.98 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
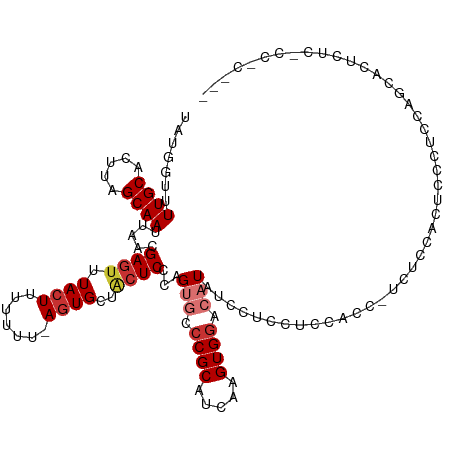
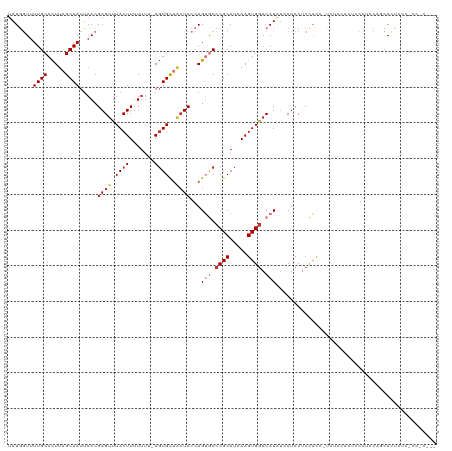
>dm3.chr2L 1768157 116 + 23011544 UAUAGUUUUGCACUUAGCAAUUAACGAGUUUACUUUUUUU-AGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCACC-UCUCCACUCCCUCCAGCACUCUCUCCACU-- ....(((((((.....))))..)))((((.((((......-))))..))))..(((((........((((((................-..)))))).......))))).........-- ( -19.93, z-score = -2.70, R) >droSim1.chr2L 1755029 117 + 22036055 UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUUUUUUUAGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCACU-CCUCCACUGCCUCCAGCACUCUCCCCACU-- ..(((..((((.....)))).....(((.............((((((.....(((((.........((((((..........))))))-....))))).....))))))))).)))..-- ( -21.53, z-score = -2.00, R) >droSec1.super_14 1702634 116 + 2068291 UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUUUUUU-AGUGCUACUCCCAGUGCCCGCAUGAAGUGACCAUAUCCUCCUCCACU-CCUCCACAGUCUCCAGCACUCUCCCCACU-- ((((((...((((((((((...((.(((....))).))..-..))))).....))))).(((.....)))))))))............-.............................-- ( -17.80, z-score = -0.84, R) >droYak2.chr2L 1741396 99 + 22324452 UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUUUUUU-AGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCAC---------UCCCUGCAGCACU----------- ..(((..((((.....)))).....((((.((((......-))))..)))))))((((..(((...((((((..........)))))---------)...))).)))).----------- ( -25.90, z-score = -3.42, R) >droEre2.scaffold_4929 1812389 107 + 26641161 UAUGCUUUUGCACUUAGCAAUUAACGAGUUUACUUUUUUU-AGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCACU-UUUCCACCACUCCCUCCACU----------- .((((....((((((((((...((.(((....))).))..-..))))).....)))))..)))).(((((((..........))))))-)...................----------- ( -20.30, z-score = -3.53, R) >dp4.chr4_group3 3150377 117 - 11692001 UAUUUUUUUGCAUUUUGCAAUUUACGAGUUUACUUUUUU--ACUGCUGCUCCCAGAACCCGCAUGAAGUGGACAUCUCCUCCUCUUCCAUCCUCAUCCUCAUCCACANCNU-UCCCCGUC .......((((.....)))).....(((((((((((...--....(((....))).........))))))))).))...................................-........ ( -12.99, z-score = -0.05, R) >droPer1.super_1 289024 118 - 10282868 UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUUUUU--AGUGCUGCUCCCAGAACCCGCAUGAAGUGGACAUCUCCUCCUCUUCCAUCCUCAUCCUCAACCACAUCCCAUCCCCGUC ...(((((((....(((((...((.(((....))).)).--..)))))....))))))).....((((.(((.......))).))))................................. ( -20.60, z-score = -1.78, R) >droWil1.scaffold_180708 640615 107 - 12563649 UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUUUUU--AGUGUCUGUCCCAGUGCCCGCAUGAAGUGGACAUG--CUCCACCAUC-CACCACCUUCCAUCUCCAUCCCC-------- .(((((.((((.....)))).....((((.((((.....--))))..((((((.(((....)))...).))))).)--))).))))).-.......................-------- ( -18.20, z-score = -0.54, R) >consensus UAUGGUUUUGCACUUAGCAAUUAACGAGUUUACUUUUUUU_AGUGCUACUCCCAGUGCCCGCAUCAAGUGGACAUAUCCUCCUCCACC_UCUCCACUCCCUCCAGCACUCUC_CC_C___ .......((((.....)))).......((((((((.......(((((......))))).......))))))))............................................... (-10.76 = -10.98 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:07 2011