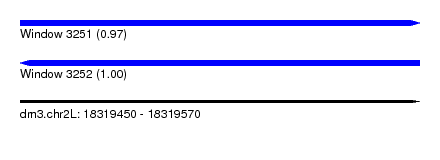
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,319,450 – 18,319,570 |
| Length | 120 |
| Max. P | 0.998865 |

| Location | 18,319,450 – 18,319,570 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.42 |
| Shannon entropy | 0.28001 |
| G+C content | 0.38595 |
| Mean single sequence MFE | -27.76 |
| Consensus MFE | -16.93 |
| Energy contribution | -18.05 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
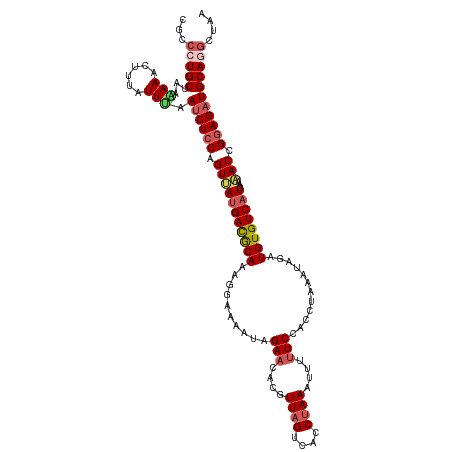
>dm3.chr2L 18319450 120 + 23011544 CACCAUGCUAAGAAACUUUAUUUGAACUCCAGUUAUGACGCAAAGGAAAAUAGAACACGUUAGUCACCUAAAUUUUUCCACCUAAAUAGAUGUGUCAUCAAAACCGGAGAUGCAGGCUAA ..((.(((((((....))))......((((.(((((((((((..(((((((((...((....))...)))...))))))..((....)).))))))))...))).))))..))))).... ( -24.20, z-score = -1.63, R) >droSim1.chr2L 18007010 120 + 22036055 CGCCAUGCUAAAAAACUUUAUUUAAUCUCCAGUUAUGACGCAAAGGAAAAUAGAACAUGUUAGUCACCCAAAAUUUUCCGCCUAAAUAGAUGUGUCAUCAAAACCGGAGAUGCAGGCUAA .(((.(((((((....))))....((((((.(((((((((((..(((((((......((.....))......)))))))..((....)).))))))))...))).))))))))))))... ( -30.60, z-score = -3.77, R) >droSec1.super_7 1946013 120 + 3727775 CGCCCUGCUAAAAAACUUUAUUUAAUCUCCAGUUAUGACGCAAAGGAAAAUAGAACAUAUUAGUCACCUAAAAUUUUCCGCCUAAAUAGAUGUGUCAUCAAAACCGGAGAUGCAGGCUUA .(((.(((((((....))))....((((((.(((((((((((..(((((((.....................)))))))..((....)).))))))))...))).))))))))))))... ( -29.90, z-score = -3.69, R) >droYak2.chr2R 4837638 120 + 21139217 CGCCCUGCUAACAAACUUAGUUGAAUCUCCAGUGACGAUGCACAGAAAAAUAGAACACGUUAGUCACCUAAAUUUUUCCACCUAAAAAUAUGUGUCAUCAACACCGGAGAUGCAGCCUGA ((..((((((((.......)))).((((((.(((..(((((((((((((((((...((....))...)))..))))))............)))).))))..))).))))))))))..)). ( -28.00, z-score = -2.91, R) >droEre2.scaffold_4845 16640844 111 - 22589142 CACCCUGCUAACAAA----GUUCGAUCUCCAGUGAUGACACAAAGAAAAGUAGA-----UUAGUCACCUAAGUUAUUCCCAAUAAAUAGAUGUGUCAUCAAAACGGAAGAUGCAGGCUGA ((.(((((.......----.....((((((..((((((((((......(((((.-----((((....)))).)))))..(........).))))))))))....)).))))))))).)). ( -26.10, z-score = -2.43, R) >consensus CGCCCUGCUAAAAAACUUUAUUUAAUCUCCAGUUAUGACGCAAAGGAAAAUAGAACACGUUAGUCACCUAAAUUUUUCCACCUAAAUAGAUGUGUCAUCAAAACCGGAGAUGCAGGCUAA ...(((((...(((......))).((((((.(((((((((((..........(((....((((....))))....)))............))))))))...))).))))))))))).... (-16.93 = -18.05 + 1.12)

| Location | 18,319,450 – 18,319,570 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.42 |
| Shannon entropy | 0.28001 |
| G+C content | 0.38595 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -27.60 |
| Energy contribution | -27.32 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.83 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.998865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
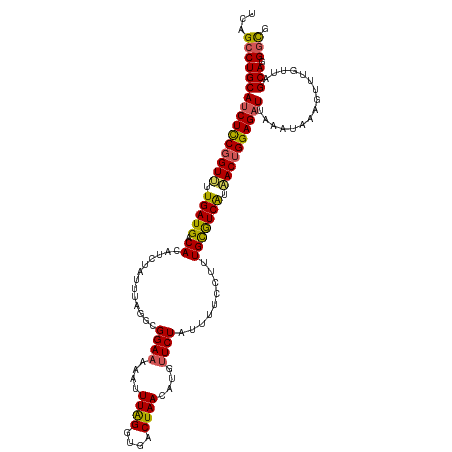
>dm3.chr2L 18319450 120 - 23011544 UUAGCCUGCAUCUCCGGUUUUGAUGACACAUCUAUUUAGGUGGAAAAAUUUAGGUGACUAACGUGUUCUAUUUUCCUUUGCGUCAUAACUGGAGUUCAAAUAAAGUUUCUUAGCAUGGUG ...((((((..((((((((.(((((.(((((((....)))))(((....((((....))))....)))..........))))))).))))))))......(((......)))))).))). ( -33.30, z-score = -2.88, R) >droSim1.chr2L 18007010 120 - 22036055 UUAGCCUGCAUCUCCGGUUUUGAUGACACAUCUAUUUAGGCGGAAAAUUUUGGGUGACUAACAUGUUCUAUUUUCCUUUGCGUCAUAACUGGAGAUUAAAUAAAGUUUUUUAGCAUGGCG ...((((((((((((((((.(((((.((..(((....))).(((((((.((((....))))........)))))))..))))))).))))))))))((((........))))))).))). ( -37.20, z-score = -4.15, R) >droSec1.super_7 1946013 120 - 3727775 UAAGCCUGCAUCUCCGGUUUUGAUGACACAUCUAUUUAGGCGGAAAAUUUUAGGUGACUAAUAUGUUCUAUUUUCCUUUGCGUCAUAACUGGAGAUUAAAUAAAGUUUUUUAGCAGGGCG ....(((((((((((((((.(((((.((..(((....))).(((((((.((((....))))........)))))))..))))))).))))))))))((((........)))))))))... ( -38.20, z-score = -4.40, R) >droYak2.chr2R 4837638 120 - 21139217 UCAGGCUGCAUCUCCGGUGUUGAUGACACAUAUUUUUAGGUGGAAAAAUUUAGGUGACUAACGUGUUCUAUUUUUCUGUGCAUCGUCACUGGAGAUUCAACUAAGUUUGUUAGCAGGGCG .....((((((((((((((.(((((.((((.......((((((((....((((....))))....))))))))...))))))))).))))))))))..(((.......))).)))).... ( -42.20, z-score = -4.40, R) >droEre2.scaffold_4845 16640844 111 + 22589142 UCAGCCUGCAUCUUCCGUUUUGAUGACACAUCUAUUUAUUGGGAAUAACUUAGGUGACUAA-----UCUACUUUUCUUUGUGUCAUCACUGGAGAUCGAAC----UUUGUUAGCAGGGUG .((.(((((((((.(((...((((((((((((((.....))))......((((....))))-----............)))))))))).)))))))..(((----...))).))))).)) ( -33.50, z-score = -3.33, R) >consensus UCAGCCUGCAUCUCCGGUUUUGAUGACACAUCUAUUUAGGCGGAAAAAUUUAGGUGACUAACAUGUUCUAUUUUCCUUUGCGUCAUAACUGGAGAUUAAAUAAAGUUUGUUAGCAGGGCG ...((((((((((((((((.(((((.((.............((((....((((....))))....)))).........))))))).))))))))))................))).))). (-27.60 = -27.32 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:48 2011