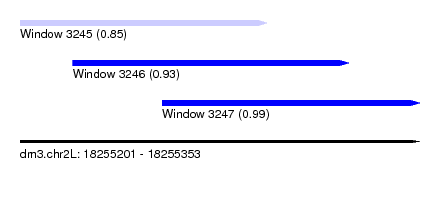
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,255,201 – 18,255,353 |
| Length | 152 |
| Max. P | 0.990245 |

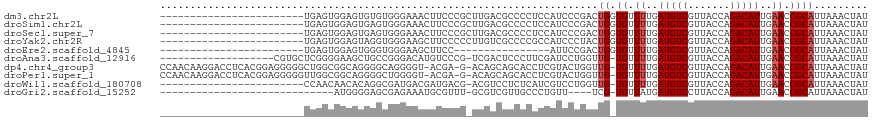
| Location | 18,255,201 – 18,255,295 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 64.64 |
| Shannon entropy | 0.67599 |
| G+C content | 0.50197 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -8.98 |
| Energy contribution | -8.75 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18255201 94 + 23011544 ------------------------UGAGUGGAGUGUGUGGGAAACUUCCCGCUUGACGCCCCUCCAUCCCGACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU ------------------------.(((.((.((((((((((....))))))...)))))))))...........(((.((..(((((.......)))))..)).))).......... ( -30.00, z-score = -2.84, R) >droSim1.chr2L 17939548 94 + 22036055 ------------------------UGAGUGGAGUGAGUGGGAAACUUCCCGCUUGACGCCCCUCCAUCCCGACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU ------------------------...((((...((((((((....))))))))((((((..((......))..))))))(..(((((.......)))))..).)))).......... ( -31.20, z-score = -3.32, R) >droSec1.super_7 1881361 94 + 3727775 ------------------------UGAGUGGAGUGAGUGGGAAACUUCCCGCUUGACGCCCCUCCAUCCCGACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU ------------------------...((((...((((((((....))))))))((((((..((......))..))))))(..(((((.......)))))..).)))).......... ( -31.20, z-score = -3.32, R) >droYak2.chr2R 4766839 94 + 21139217 ------------------------UGAGUGGAGUAGGUGGGAAGCUUCCCCCUUGUCGCCCCGCCAUCCCUACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU ------------------------..(((((.(..((((((..((..(......)..))))))))..).))))).(((.((..(((((.......)))))..)).))).......... ( -28.00, z-score = -1.78, R) >droEre2.scaffold_4845 16576022 78 - 22589142 ------------------------UGAGUGGAGUGGGUGGGAAGCUUCC----------------AUUCCGACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU ------------------------..(((((((((((.((....)))))----------------))))).))).(((.((..(((((.......)))))..)).))).......... ( -26.30, z-score = -2.50, R) >droAna3.scaffold_12916 2181453 97 + 16180835 -------------------CGUGCUCGGGGAAGCUGCCGGGACAUGUCCCG-UCGACUCCCUUCGAUCCUGGUUG-UGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU -------------------.....(((((((.(.((.(((((....)))))-.)).)))))..)))...((((((-((.((..(((((.......)))))..)).))))....)))). ( -30.40, z-score = -1.24, R) >dp4.chr4_group3 7113806 114 - 11692001 CCAACAAGGACCUCACGGAGGGGGCUGGCGGCAGGGGCAGGGGU-ACGA-G-ACAGCAGCACCUCGUACUGGUUG-UGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU ((........((((.....)))).(((.(......).)))))((-((((-(-..........)))))))((((((-((.((..(((((.......)))))..)).))))....)))). ( -32.80, z-score = 0.48, R) >droPer1.super_1 4220989 114 - 10282868 CCAACAAGGACCUCACGGAGGGGGUUGGCGGCAGGGGCUGGGGU-ACGA-G-ACAGCAGCACCUCGUACUGGUUG-UGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU (((((.....((((...))))..)))))((((....)))).(((-((((-(-..........))))))))...((-((.((..(((((.......)))))..)).))))......... ( -37.10, z-score = -0.69, R) >droWil1.scaffold_180708 11336075 92 + 12563649 ------------------------CCAACAACACAGGCGAUGACGAUGACG-ACGUCCUCUCAUCGUCCUGGUUG-UGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU ------------------------...(((((.((((((((((.((.(((.-..))).)))))))).))))))))-)(.((..(((((.......)))))..)).)............ ( -31.90, z-score = -3.83, R) >droGri2.scaffold_15252 11013770 83 - 17193109 -----------------------------AUGGGGAGCGAGAAAUGCGUUU-GCGUCGUUGCCCUGUU----UCG-UGUUAUGAUGUCCUUACCAGACAUUGAACCGCAUUAAACUAU -----------------------------..(((.(((((....(((....-)))))))).))).(((----(.(-(((..(((((((.......)))))))....)))).))))... ( -21.40, z-score = -0.91, R) >consensus ________________________UGAGUGGAGUGGGCGGGAAACUUCCCG_UCGACGCCCCUCCAUCCCGACUG_UGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAU ....................................((((..............((......))...............((..(((((.......)))))..)))))).......... ( -8.98 = -8.75 + -0.23)

| Location | 18,255,221 – 18,255,326 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.49 |
| Shannon entropy | 0.47123 |
| G+C content | 0.46521 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -14.80 |
| Energy contribution | -13.92 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.926146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18255221 105 + 23011544 ------CUUCCCGCUUGACGCCCCUCCAUCCCGACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGA--------- ------.....((.(..((((...............(((...(..(((((.......)))))..))))((((......)))).............))))..))).......--------- ( -23.30, z-score = -1.56, R) >droSim1.chr2L 17939568 105 + 22036055 ------CUUCCCGCUUGACGCCCCUCCAUCCCGACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGA--------- ------.....((.(..((((...............(((...(..(((((.......)))))..))))((((......)))).............))))..))).......--------- ( -23.30, z-score = -1.56, R) >droSec1.super_7 1881381 105 + 3727775 ------CUUCCCGCUUGACGCCCCUCCAUCCCGACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGA--------- ------.....((.(..((((...............(((...(..(((((.......)))))..))))((((......)))).............))))..))).......--------- ( -23.30, z-score = -1.56, R) >droYak2.chr2R 4766859 105 + 21139217 ------CUUCCCCCUUGUCGCCCCGCCAUCCCUACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGGCGAUGUCGA--------- ------..........((((((.(((..........(((...(..(((((.......)))))..))))((((......)))).............)))..)))))).....--------- ( -27.10, z-score = -2.57, R) >droEre2.scaffold_4845 16576042 89 - 22589142 ----------------------CUUCCAUUCCGACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGA--------- ----------------------.........((((..(((.((..(((((.......)))))..)).)))......(.(((((...........))))).).....)))).--------- ( -20.70, z-score = -1.32, R) >droAna3.scaffold_12916 2181478 103 + 16180835 -------UGUCCCGUCGACUCCCUUCGAUCCUGGUUG-UGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGA--------- -------.....(((((((..((.........)).((-(((((..(((((.......)))))..))..((((......))))......)))))....))))))).......--------- ( -26.20, z-score = -1.63, R) >dp4.chr4_group3 7113841 110 - 11692001 GGCAGGGGUACGAGACAGCAGCACCUCGUACUGGUUG-UGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUCGACUACGACGA--------- .((((.((((((((..........))))))))...((-(((((..(((((.......)))))..))..((((......))))......)))))))))((((....))))..--------- ( -34.80, z-score = -2.35, R) >droPer1.super_1 4221024 110 - 10282868 GGCUGGGGUACGAGACAGCAGCACCUCGUACUGGUUG-UGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUCGACGACGACGA--------- .(((((((((((((..........))))))))...((-((.((..(((((.......)))))..)).)))).....))).))..............(((((....))))).--------- ( -33.30, z-score = -1.55, R) >droWil1.scaffold_180708 11336103 95 + 12563649 ---------------CGUCCUCUCAUCGUCCUGGUUG-UGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUCGAGGAUGUCGA--------- ---------------(((((((....(((..((..((-(((((..(((((.......)))))..)...((((......))))...))))))))..)))..)))))))....--------- ( -27.20, z-score = -2.00, R) >droVir3.scaffold_12963 2056265 93 - 20206255 ------------------CGCGUCAUUGCCCUGCGUCGUGUUUUGAUGUCCUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGGCGACGA--------- ------------------..((((...(((..((((.((((((..(((((.......)))))..))..((((......))))........)))).))))....))))))).--------- ( -26.40, z-score = -1.79, R) >droGri2.scaffold_15252 11013785 108 - 17193109 ------------UGCGUUUGCGUCGUUGCCCUGUUUCGUGUUAUGAUGUCCUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGAUCCUUGCGUCGCGUUCUCGUC ------------.(((..((((..(((((..(((...(((((..((((((.......)))))).....((((......))))...))))))))..)))))...)))).)))......... ( -25.00, z-score = -0.80, R) >droMoj3.scaffold_6500 5487058 93 - 32352404 ------------------CGCGUCGUUGCCCUGUGUCGUGUUUUGAUGUCCUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGAUGACGGCGACGA--------- ------------------(((((((((((..(((((...(.((..(((((.......)))))..)).)((((......))))......)))))..)))))))).)))....--------- ( -32.30, z-score = -3.67, R) >consensus ________U_C___UCGACGCCCCUCCAUCCUGACUGGUGUUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGA_________ ..........................((((.........(.((..(((((.......)))))..)).)((((......)))).....................))))............. (-14.80 = -13.92 + -0.88)

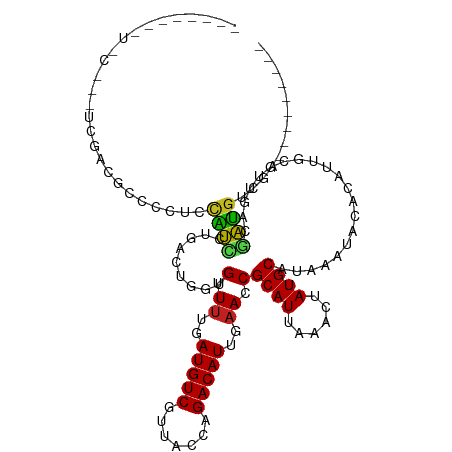
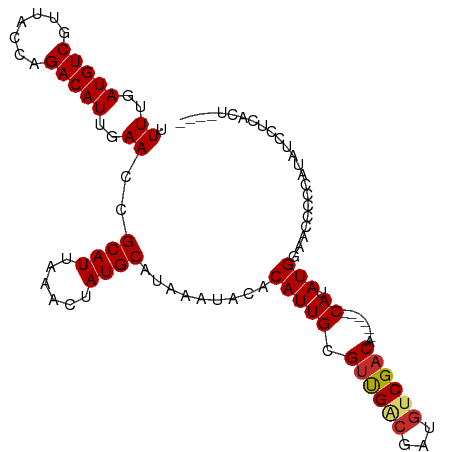
| Location | 18,255,255 – 18,255,353 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.48 |
| Shannon entropy | 0.29247 |
| G+C content | 0.42812 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.81 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18255255 98 + 23011544 UUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGACA------CAUAUGGAACCCCCAUAUCCUCACU---- .((..(((((.......)))))..))..((((......))))............((.((((((...)))))).------))(((((.....)))))........---- ( -22.80, z-score = -3.11, R) >droSim1.chr2L 17939602 98 + 22036055 UUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGACA------CAUAUGGAACCCCCAUAUCCUCACU---- .((..(((((.......)))))..))..((((......))))............((.((((((...)))))).------))(((((.....)))))........---- ( -22.80, z-score = -3.11, R) >droSec1.super_7 1881415 98 + 3727775 UUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGACA------CAUAUGGAACCCCCAUAUCCUCACU---- .((..(((((.......)))))..))..((((......))))............((.((((((...)))))).------))(((((.....)))))........---- ( -22.80, z-score = -3.11, R) >droYak2.chr2R 4766893 98 + 21139217 UUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGGCGAUGUCGACA------CAUAUGGAACCCCCAUAUCCUCACU---- .((..(((((.......)))))..))..((((......))))........(((((((....))))))).((..------.((((((.....))))))..))...---- ( -25.00, z-score = -3.08, R) >droEre2.scaffold_4845 16576060 98 - 22589142 UUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGACA------CAUAUGGAACCCCCAUAUCCUCACU---- .((..(((((.......)))))..))..((((......))))............((.((((((...)))))).------))(((((.....)))))........---- ( -22.80, z-score = -3.11, R) >droAna3.scaffold_12916 2181510 86 + 16180835 UUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGACA------CAUAUGGAACCUC---------------- .((..(((((.......)))))..))((((((......))))............((.((((((...)))))).------))...))......---------------- ( -20.70, z-score = -2.45, R) >droWil1.scaffold_180708 11336127 96 + 12563649 UUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUCGAGGAUGUCGACA------CAUAUGGCCCCCAAAGGCACUCG------ .((..(((((.......)))))..))..((((......)))).........(((((.(((((.....))))).------)).)))(((......))).....------ ( -24.70, z-score = -2.21, R) >droVir3.scaffold_12963 2056287 94 - 20206255 UUUUGAUGUCCUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGGCGACGACAUCGUCACAUAUGCGCCCCCCC-------------- .((..(((((.......)))))..))..((((......)))).............((((.....(.((((....)))).)....))))......-------------- ( -22.10, z-score = -2.86, R) >droMoj3.scaffold_6500 5487080 108 - 32352404 UUUUGAUGUCCUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGAUGACGGCGACGACAUCAUCGCAUAUGCCCCCGCCCGUCUCUGUCACCGC .((..(((((.......)))))..))..((((......)))).............(((.((((((.((((........((....)).......)))).)))))).))) ( -28.96, z-score = -3.42, R) >consensus UUUUGAUGUCGUUACCAGACAUUGAACCGCAUUAAACUAUGCAUAAAUACACAUUGCGUUGACGAUGUCGACA______CAUAUGGAACCCCCAUAUCCUCACU____ .((..(((((.......)))))..))((((((......))))...............((((((...))))))............))...................... (-16.32 = -16.81 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:44 2011