| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,239,555 – 18,239,647 |
| Length | 92 |
| Max. P | 0.712171 |

| Location | 18,239,555 – 18,239,647 |
|---|---|
| Length | 92 |
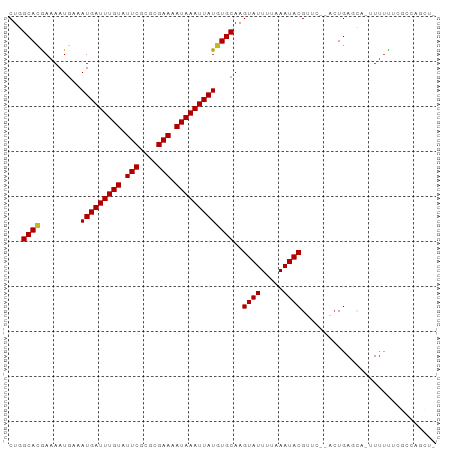
| Sequences | 12 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 84.61 |
| Shannon entropy | 0.33817 |
| G+C content | 0.31877 |
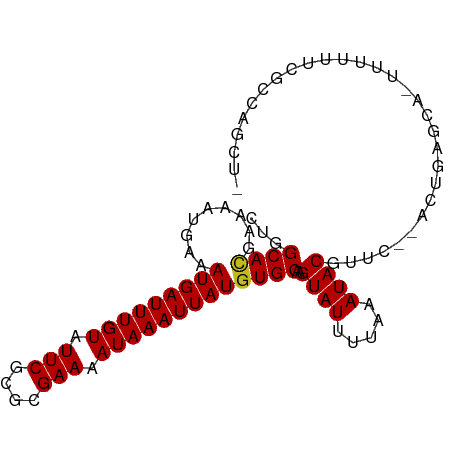
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.31 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.712171 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 18239555 92 - 23011544 CUGGCACGAAAAUGAAAUGAUUUGUAUUCGCGCGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGUUC--ACUGAGCGUUUUUUUCGCCAGCU- (((((..(((((((..(((((((((.(((....))).)))))))))(((...((((.....))))...)--))....)))))))...)))))..- ( -22.90, z-score = -1.57, R) >droSim1.chr2L 17922174 91 - 22036055 CUGGCACGAAAAUGAAAUGAUUUGUAUUCGCGCGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGUUC--ACUGAGCA-UUUUUUCGCCAGCU- (((((..(((((((..(((((((((.(((....))).)))))))))(((...((((.....))))...)--))....))-)))))..)))))..- ( -23.90, z-score = -2.29, R) >droSec1.super_7 1865501 91 - 3727775 CUGGCAUAAAAAUGAAAUGAUUUGUAUUCGCGCGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGUUC--ACUGAGCA-UUUUUUCGCCAGCU- (((((..(((((((..(((((((((.(((....))).)))))))))(((...((((.....))))...)--))....))-)))))..)))))..- ( -24.40, z-score = -2.85, R) >droYak2.chr2R 4750052 91 - 21139217 CUGGCACGAAAAUGAAAUGAUUUGUAUUCGUGUGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGUUC--ACUGAGCA-UUUUUUCGCCAGCU- (((((..(((((((..(((((((((.(((....))).)))))))))(((...((((.....))))...)--))....))-)))))..)))))..- ( -23.00, z-score = -2.49, R) >droEre2.scaffold_4845 16560298 91 + 22589142 CUGGCACGAAAAUGAAAUGAUUUGUAUUCGCGCGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGUUC--ACUGAGCA-UUUUUUCGCCAGCU- (((((..(((((((..(((((((((.(((....))).)))))))))(((...((((.....))))...)--))....))-)))))..)))))..- ( -23.90, z-score = -2.29, R) >droAna3.scaffold_12916 2166463 91 - 16180835 CUGGCACGAAAAUGAAAUGAUUUGUAUUCGCGCGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGUUC--ACUGAGCAUUUUUCUCGGAACU-- ((((...(((((((..(((((((((.(((....))).)))))))))(((...((((.....))))...)--))....))))))).))))....-- ( -17.00, z-score = -0.43, R) >dp4.chr4_group3 7095935 94 + 11692001 CUUGCACGAAAAUGAAAUGAUUUGUAUUCGCGCGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGUUC-UACUGAGCACAUUUUUUCCCGUGUU ...(((((........(((((((((.(((....))).)))))))))((((..((((.....))))..((-....)))))).........))))). ( -20.20, z-score = -1.70, R) >droPer1.super_1 4201634 94 + 10282868 CUUGCACGAAAAUGAAAUGAUUUGUAUUCGCGCGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGUUC-UACUGAGCACAUUUUUUCCCGUGUU ...(((((........(((((((((.(((....))).)))))))))((((..((((.....))))..((-....)))))).........))))). ( -20.20, z-score = -1.70, R) >droWil1.scaffold_180708 11318450 87 - 12563649 CUUGCAAGAAAAUGAAAUGAUUUGUAUUCGCGCGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGCGCAAUGUGAAAAAUUCUUUU-------- .(..((..........(((((((((.(((....))).)))))))))((((..((((.....))))))))..))..)...........-------- ( -15.10, z-score = 0.29, R) >droMoj3.scaffold_6500 26840346 83 - 32352404 CUUGCACGAAAAUGAAAUGAUUUGUAUUCGCGAGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGAAU--UCUGACUUCGUUUU---------- (((((((.........(((((((((.(((....))).))))))))))))))))..........(((((.--......)))))...---------- ( -19.60, z-score = -2.60, R) >droVir3.scaffold_12963 18125977 83 - 20206255 CUUGCACGAAAAUGAAAUGAUUUGUAUUCGCGAGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGAAU--UCUGACUUUUUUCU---------- (((((((.........(((((((((.(((....))).))))))))))))))))................--..............---------- ( -17.50, z-score = -2.36, R) >droGri2.scaffold_15252 6158604 84 - 17193109 CUUGCACGAAAAUUAAAUGAUUUGUAUUCGUGAGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACG-ACUUUCUGAAACUUUUUU---------- (((((((.........(((((((((.(((....))).)))))))))))))))).............-..................---------- ( -18.00, z-score = -3.10, R) >consensus CUGGCACGAAAAUGAAAUGAUUUGUAUUCGCGCGAAAAUAAAUUAUGUGCAAGUAUUUUAAAUACGUUC__ACUGAGCA_UUUUUUCGCCAGCU_ ...((((.........(((((((((.(((....))).)))))))))))))..((((.....)))).............................. (-12.29 = -12.31 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:41 2011