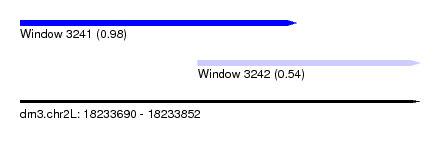
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,233,690 – 18,233,852 |
| Length | 162 |
| Max. P | 0.983734 |

| Location | 18,233,690 – 18,233,802 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Shannon entropy | 0.37450 |
| G+C content | 0.41827 |
| Mean single sequence MFE | -30.29 |
| Consensus MFE | -20.39 |
| Energy contribution | -21.28 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
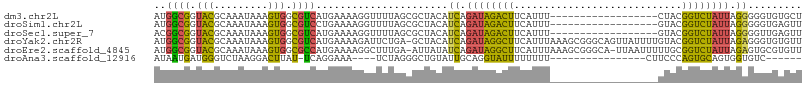
>dm3.chr2L 18233690 112 + 23011544 CUGAUGAUGAUGACGGGGCUGUUACUGUCAUUAAAGUUGGACCAUGAAUGUCUAACGGAUAAAGAGAUUCAAAUGGCGGUACGCAAAUAAAGUGGCGUCAUGAAAAGGUUUU (..((..(((((((((........)))))))))..))..)(((.(((((.(((.........))).))))).((((((.(((.........))).)))))).....)))... ( -33.70, z-score = -3.69, R) >droSim1.chr2L 17916206 112 + 22036055 CUGAUGAUGAUGACGGGGCUGUUACUGUCAUUAAAGUUGGACCAUGAAUGUCUAACGCAUAAAGAGAUUCAAAUGGCGGUACGCAAAUAAAGUGGCGUCCUGAAAAGGUUUU (..((..(((((((((........)))))))))..))..)(((.(((((.(((.........))).)))))...((((.(((.........))).)))).......)))... ( -30.80, z-score = -2.15, R) >droSec1.super_7 1859436 112 + 3727775 CUGAUGAUGAUGACGGGGCUGUUACUGUCAUUAAAGUUGGACCAUGAAUGUCUAACGCAUAAAGAGAUUCAAACGGCGGUACGCAAAUAAAGUGGCGUCAUGAAAAGGUUUU (..((..(((((((((........)))))))))..))..)(((.(((((.(((.........))).)))))...((((.(((.........))).)))).......)))... ( -32.00, z-score = -2.66, R) >droYak2.chr2R 4743947 112 + 21139217 CUGAUGAUGAUGACGGGGCUGCUACUGUCAUUAAAGUUGGACCAUGAAUGUCUAACGCAUAAAGAGAUUCAAAUGGCGGUACGCAAAUAAAGUGGCGUCAUGAAAAGAUUCU ...(((.(((((((((........)))))))))..(((((((.......))))))).)))....(((.((..((((((.(((.........))).)))))).....)).))) ( -31.00, z-score = -2.41, R) >droEre2.scaffold_4845 16552993 112 - 22589142 CUGAUGACGAUGAGGGGGCUGUUACUGUCAUUAAAGUUGGACCGUGAACGUCUAACGCAUAAAGAGAUUCAAAUGGCGGUACGCAAAUAAAGUGGCGCCAUGAAAAGGCUUU .((((((((.(((.(....).))).))))))))..(((((((.(....))))))))......((((......((((((.(((.........))).)))))).......)))) ( -31.82, z-score = -1.91, R) >droAna3.scaffold_12916 2160567 91 + 16180835 ---------AUG--GGGCCUGUUACCGUCAUUAAAGUUAGUCGGGAAAUG---AACACAUUACGAAGU--AAAUAAUGAUGGGUC--UAAGGACUUAUUCAGGAAAUCU--- ---------..(--((.((((...(((((((((...(((.(((...((((---....)))).)))..)--)).)))))))))(((--....))).....))))...)))--- ( -22.40, z-score = -1.71, R) >consensus CUGAUGAUGAUGACGGGGCUGUUACUGUCAUUAAAGUUGGACCAUGAAUGUCUAACGCAUAAAGAGAUUCAAAUGGCGGUACGCAAAUAAAGUGGCGUCAUGAAAAGGUUUU .......(((((((((........)))))))))..(((((((.......)))))))..................((((.(((.........))).))))............. (-20.39 = -21.28 + 0.89)

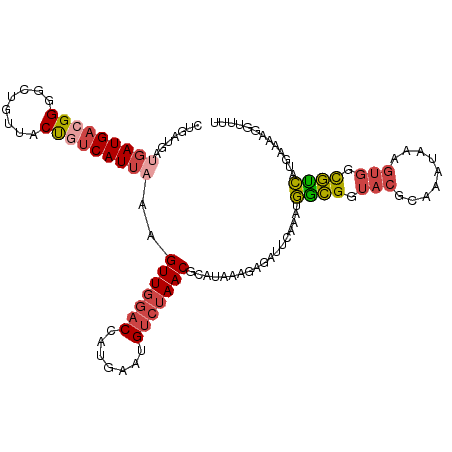
| Location | 18,233,762 – 18,233,852 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 66.16 |
| Shannon entropy | 0.59883 |
| G+C content | 0.42598 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -11.83 |
| Energy contribution | -10.81 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.535421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18233762 90 + 23011544 AUGGCGGUACGCAAAUAAAGUGGCGUCAUGAAAAGGUUUUAGCGCUACAUCAGAUAGACUUCAUUU------------------CUACGGUCUAUUAGGGGGUGUGCU .....(((((((.......(((((((..((((.....))))))))))).((.((((((((......------------------....)))))))).))..))))))) ( -25.30, z-score = -2.04, R) >droSim1.chr2L 17916278 90 + 22036055 AUGGCGGUACGCAAAUAAAGUGGCGUCCUGAAAAGGUUUUAGCGCUACAUCAGAUAGACUUCAUUU------------------GUACGGUCUAUUAGGGGGUGAGUU ..(((..(((.(.......((((((((((....))).....))))))).((.((((((((......------------------....)))))))).))).))).))) ( -24.30, z-score = -1.49, R) >droSec1.super_7 1859508 90 + 3727775 ACGGCGGUACGCAAAUAAAGUGGCGUCAUGAAAAGGUUUUAGCGCUACAUCAGAUAGACUUCAUUU------------------GUACGGUCUAUUAGGGGUUGAGUU .((((..............(((((((..((((.....))))))))))).((.((((((((......------------------....)))))))).)).)))).... ( -19.80, z-score = -0.46, R) >droYak2.chr2R 4744019 107 + 21139217 AUGGCGGUACGCAAAUAAAGUGGCGUCAUGAAAAGAUUCUGA-GCUACAUCAGAUAGGCUUCAUUUAAAGCGGGCAGUUAUUUUGUACGGUCUAUUAGAGGGUGUGUU ..(((.(((((.((((((.(((((.(((.(((....))))))-))))).........((((......))))......))))))))))).)))................ ( -24.80, z-score = -0.42, R) >droEre2.scaffold_4845 16553065 106 - 22589142 AUGGCGGUACGCAAAUAAAGUGGCGCCAUGAAAAGGCUUUGA-AUUAUAUCAGAUAGGCUUCAUUUAAAGCGGGCA-UUAAUUUUUGCGGUCUAUUAGAGUGCGUGUU ((((((.(((.........))).))))))......(((((((-((.(..((.....))..).))))))))).....-.........(((.((.....)).)))..... ( -25.50, z-score = -0.15, R) >droAna3.scaffold_12916 2160623 81 + 16180835 AUAAUGAUGGGUCUAAGGACUUAU-UCAGGAAA----UCUAGGGCUGUAUUGCAGGUAUUUUUUUU----------------CUUCCCAGUGCAGUGGUGUC------ ....(((((((((....)))))).-))).....----......(((((((((..((..........----------------...)))))))))))......------ ( -16.92, z-score = 0.18, R) >consensus AUGGCGGUACGCAAAUAAAGUGGCGUCAUGAAAAGGUUUUAGCGCUACAUCAGAUAGACUUCAUUU__________________GUACGGUCUAUUAGGGGGUGUGUU ..((((.(((.........))).))))......................((.((((((((............................)))))))).))......... (-11.83 = -10.81 + -1.02)
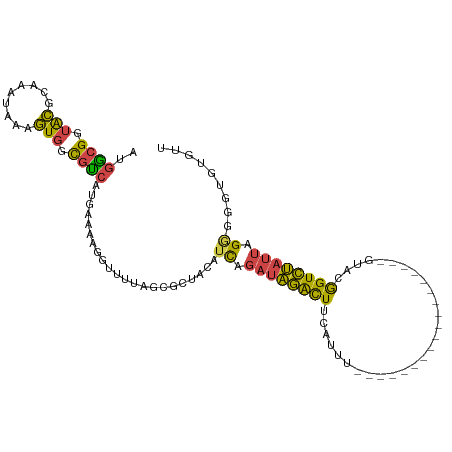
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:40 2011