
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,233,497 – 18,233,600 |
| Length | 103 |
| Max. P | 0.576368 |

| Location | 18,233,497 – 18,233,600 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.97 |
| Shannon entropy | 0.42069 |
| G+C content | 0.39883 |
| Mean single sequence MFE | -27.91 |
| Consensus MFE | -13.56 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.557853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
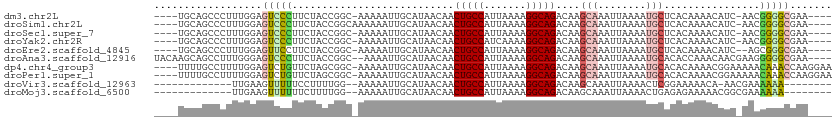
>dm3.chr2L 18233497 103 + 23011544 ----UUCGCCCCGUU-GAUGUUUUGUGAGCAUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUUU-GCCGGUAGAAGGGACUCCAAAGGGCUGCA---- ----...((((..((-(..(((..(..((((........))))..)...(((((((.((((((..(((....)))..))-)))).))))))))))..))).))))....---- ( -32.40, z-score = -1.87, R) >droSim1.chr2L 17916012 104 + 22036055 ----UUCGCCCCGUU-GAUGUUUUGUGAGCAUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUUUUGCCGGUAGAAGGGACUCCAAAGGGCUGCA---- ----...((((..((-(..(((..(..((((........))))..)...(((((((.(((((((((.....))))....))))).))))))))))..))).))))....---- ( -32.00, z-score = -1.78, R) >droSec1.super_7 1859243 103 + 3727775 ----UUCGCCCCGUU-GAUGUUUUGUGAGCAUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUUU-GCCGGUAGAAGGGACUCCAAAGGGCUGCA---- ----...((((..((-(..(((..(..((((........))))..)...(((((((.((((((..(((....)))..))-)))).))))))))))..))).))))....---- ( -32.40, z-score = -1.87, R) >droYak2.chr2R 4743754 103 + 21139217 ----UUCGCCCCGUU-GAUGUUUUGUGAGCAUUUUAAUUUGCUUGUCUGCCUUUUGAUGGCAGUUGUUAUGCAAUUUUU-GCCGGUAGAAGGGACUCCAAAGGGCUGCA---- ----...((((..((-(..(((..(..((((........))))..)...(((((((.((((((..(((....)))..))-)))).))))))))))..))).))))....---- ( -32.10, z-score = -1.62, R) >droEre2.scaffold_4845 16552802 102 - 22589142 ----UUCGCCCGCU--GAUGUUUUGUGAGCAUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUUU-GCCGGUAGAAGGAACUCCAAAGGGCUGCA---- ----...((((..(--(..(((..(..((((........))))..)...(((((((.((((((..(((....)))..))-)))).))))))))))..))..))))....---- ( -32.40, z-score = -1.96, R) >droAna3.scaffold_12916 2160362 107 + 16180835 ----UUCGCCCCCUUCGUUGUUUGGUGUGCAUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUU--GCCGGUAGAAGGGACUCCCAAAGGCUGCUUGUA ----...(((((((((.....(((((.(((((..(((.........(((((.......))))))))..))))).....--)))))..)))))).........)))........ ( -27.70, z-score = -0.44, R) >dp4.chr4_group3 7088247 108 - 11692001 UUCCUUGGUUUGUUUUUCCGUUUUGUGUGCAUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUUU-GCCGCUAGAACAGACUCCAAAAGGCAAAA---- ..((((((((((((((........(((.(((....((.((((.((.(((((.......)))))....)).)))).)).)-))))).))))))))))....)))).....---- ( -26.80, z-score = -1.71, R) >droPer1.super_1 4193980 108 - 10282868 UUCCUUGGUUUGUUUUUCCGUUUUGUGUGCAUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUUU-GCCGCUAGAACAGACUCCAAAAGGCAAAA---- ..((((((((((((((........(((.(((....((.((((.((.(((((.......)))))....)).)))).)).)-))))).))))))))))....)))).....---- ( -26.80, z-score = -1.71, R) >droVir3.scaffold_12963 18117466 89 + 20206255 --------UUUUUUCGUU-UGUUUUUCCGAGUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUUU--CCAAAAGGAAAAACUUCAA------------- --------..........-.((((((((.......((.((((.((.(((((.......)))))....)).)))).))..--......)))))))).....------------- ( -20.76, z-score = -2.98, R) >droMoj3.scaffold_6500 26830876 90 + 32352404 --------UUUUUUCGCCGUUUUUCUCUCAGUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUUU--CCAAAAGAAAAAACUUCAA------------- --------..........((((((.(((.......((.((((.((.(((((.......)))))....)).)))).))..--.....))))))))).....------------- ( -15.74, z-score = -1.78, R) >consensus ____UUCGCCCCGUU_GAUGUUUUGUGAGCAUUUUAAUUUGCUUGUCUGCCUUUUAAUGGCAGUUGUUAUGCAAUUUUU_GCCGGUAGAAGGGACUCCAAAGGGCUGCA____ .......((((........(((((..............((((.((.(((((.......)))))....)).))))................)))))......))))........ (-13.56 = -13.92 + 0.36)



| Location | 18,233,497 – 18,233,600 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Shannon entropy | 0.42069 |
| G+C content | 0.39883 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -13.68 |
| Energy contribution | -13.21 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18233497 103 - 23011544 ----UGCAGCCCUUUGGAGUCCCUUCUACCGGC-AAAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAAUGCUCACAAAACAUC-AACGGGGCGAA---- ----....(((((((((.(((((((.((..(((-(....(((.....))).))))..)).))))..))).((((........)))).........))-)).)))))...---- ( -23.60, z-score = -0.94, R) >droSim1.chr2L 17916012 104 - 22036055 ----UGCAGCCCUUUGGAGUCCCUUCUACCGGCAAAAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAAUGCUCACAAAACAUC-AACGGGGCGAA---- ----....((((((((..((...........((((....)))).......(((((.......)))))...((((........))))......))..)-)).)))))...---- ( -24.50, z-score = -1.31, R) >droSec1.super_7 1859243 103 - 3727775 ----UGCAGCCCUUUGGAGUCCCUUCUACCGGC-AAAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAAUGCUCACAAAACAUC-AACGGGGCGAA---- ----....(((((((((.(((((((.((..(((-(....(((.....))).))))..)).))))..))).((((........)))).........))-)).)))))...---- ( -23.60, z-score = -0.94, R) >droYak2.chr2R 4743754 103 - 21139217 ----UGCAGCCCUUUGGAGUCCCUUCUACCGGC-AAAAAUUGCAUAACAACUGCCAUCAAAAGGCAGACAAGCAAAUUAAAAUGCUCACAAAACAUC-AACGGGGCGAA---- ----....((((((((..((...........((-(.....))).......(((((.......)))))...((((........))))......))..)-)).)))))...---- ( -23.60, z-score = -0.96, R) >droEre2.scaffold_4845 16552802 102 + 22589142 ----UGCAGCCCUUUGGAGUUCCUUCUACCGGC-AAAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAAUGCUCACAAAACAUC--AGCGGGCGAA---- ----....((((.(((..(((..........((-(.....))).......(((((.......)))))...((((........)))).....)))..)--)).))))...---- ( -24.20, z-score = -1.04, R) >droAna3.scaffold_12916 2160362 107 - 16180835 UACAAGCAGCCUUUGGGAGUCCCUUCUACCGGC--AAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAAUGCACACCAAACAACGAAGGGGGCGAA---- .....((..((....))..(((((((.....((--(....))).......(((((.......)))))....(((........)))............)))))))))...---- ( -25.80, z-score = -1.22, R) >dp4.chr4_group3 7088247 108 + 11692001 ----UUUUGCCUUUUGGAGUCUGUUCUAGCGGC-AAAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAAUGCACACAAAACGGAAAAACAAACCAAGGAA ----.....(((((((...((((((......((-(.....))).......(((((.......)))))....(((........)))......))))))....)))...)))).. ( -21.20, z-score = -0.69, R) >droPer1.super_1 4193980 108 + 10282868 ----UUUUGCCUUUUGGAGUCUGUUCUAGCGGC-AAAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAAUGCACACAAAACGGAAAAACAAACCAAGGAA ----.....(((((((...((((((......((-(.....))).......(((((.......)))))....(((........)))......))))))....)))...)))).. ( -21.20, z-score = -0.69, R) >droVir3.scaffold_12963 18117466 89 - 20206255 -------------UUGAAGUUUUUCCUUUUGG--AAAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAACUCGGAAAAACA-AACGAAAAAA-------- -------------.....((((((((.(((..--..)))((((.......(((((.......)))))....))))..........)))))))).-..........-------- ( -19.10, z-score = -3.07, R) >droMoj3.scaffold_6500 26830876 90 - 32352404 -------------UUGAAGUUUUUUCUUUUGG--AAAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAACUGAGAGAAAAACGGCGAAAAAA-------- -------------(((..((((((.((((..(--.....((((.......(((((.......)))))....)))).......)..))))))))))..))).....-------- ( -19.80, z-score = -2.66, R) >consensus ____UGCAGCCCUUUGGAGUCCCUUCUACCGGC_AAAAAUUGCAUAACAACUGCCAUUAAAAGGCAGACAAGCAAAUUAAAAUGCUCACAAAACAUC_AAAGGGGCGAA____ ..................(((((...........................(((((.......)))))....(((........)))................)))))....... (-13.68 = -13.21 + -0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:38 2011