
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,221,759 – 18,221,878 |
| Length | 119 |
| Max. P | 0.854114 |

| Location | 18,221,759 – 18,221,878 |
|---|---|
| Length | 119 |
| Sequences | 7 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 59.74 |
| Shannon entropy | 0.73747 |
| G+C content | 0.45428 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -10.12 |
| Energy contribution | -9.07 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
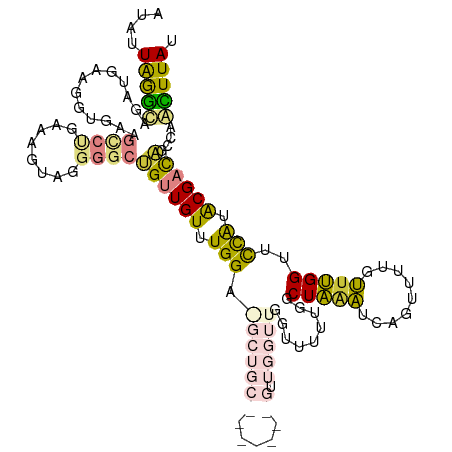
>dm3.chr2L 18221759 119 - 23011544 AUAUUGGGCAGAUAAAGGUGAUGCCCGAAAGUAGGGGCUGUUGUUUGGAGGCUGCGGCAUACGUUGGUUGGUUUUUGGCUAAAUCUUUUUUGUCUGGUUCCAUACGACAACCAGCUUAU ...(((((((...........)))))))((((.((...((((((.(((((.(...((((....(((((..(...)..)))))........)))).).))))).)))))).)).)))).. ( -35.90, z-score = -1.00, R) >droSim1.chr2L 17904281 118 - 22036055 AUAUUAGGCAGAUGAAGGGGAUGCCUAAAAGUAGGGGCUGUUGUUUGGAGGCUGCGGCAUACGUUGGUUGGUUUUUGGCUAAAUCAGUUUUUUUUGGUUCCAUACGACAGCU-GCUUAU ...(((((((...........)))))))....((.(((((((((.(((((.(..((((....)))).(((((((......)))))))........).))))).)))))))))-.))... ( -37.30, z-score = -2.17, R) >droSec1.super_7 1847506 119 - 3727775 AUAUUAGGCAGAUGAAGGGGAUGCCCAAAAGUAGGGGCUGUUGUUUGGAGGCUGCGGCAUACGUUGGUUGGUUUUUGGCUAAAUCAGUUUUUUUUGGUUCCAUACGACAGCUAGCUUAU ....(((((.......(((....))).........(((((((((.(((((.(..((((....)))).(((((((......)))))))........).))))).))))))))).))))). ( -36.10, z-score = -1.52, R) >droYak2.chr2R 4727890 118 - 21139217 AUAUUAGGCAGAUGAAGGUGAAGUCUGCAAAUAAGGGCUGUUGUUUGGAAGCUGCCUAUGUUGUUGGUUGGAAUUUGGCUAGAACAGUUUU-UUUGGUACCAAGCGACUGCCAAUUUAU .......((((((.........)))))).......(((.(((((((((....((((..((((.(((((..(...)..))))))))).....-...))))))))))))).)))....... ( -33.70, z-score = -1.40, R) >droEre2.scaffold_4845 16541404 90 + 22589142 AUAUUAGGCAGACGAAGGCGAAGUCUGCAAAU-GGGGCUGUUGUUUGGA---------------------------UCCUAAAGCUGUUUU-UUUGGUACUGAACGACAGCCAACUUAU .......((((((.........))))))....-..((((((((((..(.---------------------------..((((((......)-)))))..)..))))))))))....... ( -31.80, z-score = -3.72, R) >droPer1.super_1 4179503 94 + 10282868 AUACUGAAUAAAAGGUGGUGGAAA--GAGGGU-GGGAAAGCUGUUUGGU----------------------UUUCCGCCUGAGCAGGCAUCGCUCGGCCUCACGCGCUUGACAAUUUAU ....(((((...(((((((((...--..((((-(((((((((....)))----------------------)))))((((....))))..)))))....)))).)))))....))))). ( -31.10, z-score = -1.37, R) >dp4.chr4_group3 7073321 94 + 11692001 AUACUGAAUAAAAGGUGGUGGAAC--GAGGGU-GGGAAAGCUGUUUGGU----------------------UUUCCGCCUGAGCAGGCAUCGCUCGGCCUCAUGCGCUUGACAAUUUAU ....(((((...(((((((((..(--((((((-.((((((((....)))----------------------)))))((((....))))))).))))..).))).)))))....))))). ( -30.70, z-score = -1.26, R) >consensus AUAUUAGGCAGAUGAAGGUGAAGCCUGAAAGUAGGGGCUGUUGUUUGGA_GCUGC_______GUUGGUUGGUUUUUGGCUAAAUCAGUUUUGUUUGGUUCCAUACGACAGCCAACUUAU ....(((((.............((((........))))((((((.(((.....(((.....)))..............(((((.........)))))..))).))))))....))))). (-10.12 = -9.07 + -1.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:35 2011