
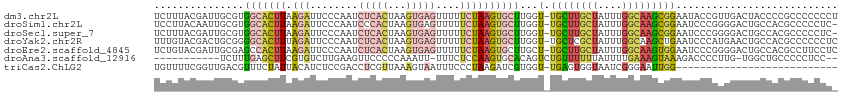
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,192,371 – 18,192,484 |
| Length | 113 |
| Max. P | 0.893458 |

| Location | 18,192,371 – 18,192,484 |
|---|---|
| Length | 113 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.87 |
| Shannon entropy | 0.73189 |
| G+C content | 0.47602 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -10.01 |
| Energy contribution | -11.20 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
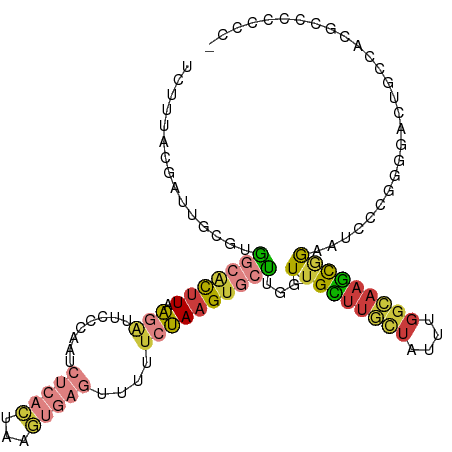
>dm3.chr2L 18192371 113 + 23011544 UCUUUACGAUUGCGUGGCACUUAAGAUUCCCAAUCUCACUAAGUGAGUUUUUCUAAGUGCUUGGU-UGCUUGCUAUUUGGCAAGCGGAAUACCGUUGACUACCCCGCCCCCCCU ...........(((.((((((((.((........(((((...)))))....))))))))))((((-((((((((....)))))))((....))...)))))...)))....... ( -30.60, z-score = -2.23, R) >droSim1.chr2L 17871125 112 + 22036055 UCCUUACAAUUGCGUGGCACUUAAGAUUCCCAAUCCCACUAAGUGAGUUUUUCUAAGUGCUUGGU-UGCUUGCUAUUUGGCAAGCGGAAUCCCGGGGACUGCCACGCCCCCUC- ...........((((((((.....((((((.......((((((((..(......)..))))))))-.(((((((....)))))))))))))..(....)))))))))......- ( -40.20, z-score = -3.38, R) >droSec1.super_7 1818010 112 + 3727775 UCUUUACGAUUGCGUGGCACUUAAGAUUCCCAAUCUCACUAAGUGAGUUUUUCUAAGUGCUUGGU-UGCUUGCUAUUUGGCAAGCGGAAUCCCGGGGACUGCCACGCCCCCUC- ...........((((((((.....((((((.......((((((((..(......)..))))))))-.(((((((....)))))))))))))..(....)))))))))......- ( -40.20, z-score = -3.35, R) >droYak2.chr2R 4698071 113 + 21139217 UUUGUACGACUGCGGGGCACUUUAGAUUCCCAAUCUCACUAAGUGAGUUUUUCUAAGUGCUUGGU-UGCUCGCUAUUUGGCAAGCUGAAUCCCAUGAACUGCCACGCCCCCCUC .............(((((.......................(((((((....((((....)))).-.)))))))...(((((..(((.....)).)...))))).))))).... ( -28.10, z-score = -0.40, R) >droEre2.scaffold_4845 16511449 113 - 22589142 UCUGUACGAUUGCGAGCCACUUAAGAUUCCCAAUCUCACUAAGUGAGUUUUUCUAAGUGCUUGCU-UGCUUGCUAUUUGGCAAGUGGAAUCCCGGGGACUGCCACGCCUUCCUC .......((((((((((.(((((.((........(((((...)))))....)))))))))))))(-..((((((....))))))..)))))..(((((..((...))..))))) ( -34.20, z-score = -1.99, R) >droAna3.scaffold_12916 2120625 99 + 16180835 -----------UCUUUGAGCUUCGUGUCUUGAAGUUCCCCCAAAUU-UUUCUCCAAGUGCACAGUCUGUUUUUUAUUUUGAAAGUAAAGACCCCUUG-UGGCUGCCCCCUCC-- -----------.....((((((((.....)))))))).........-........(((.((((((((..(((((.....)))))...))))....))-))))).........-- ( -17.10, z-score = -1.24, R) >triCas2.ChLG2 149964 86 - 12900155 UGUUUUCGGUUGACGUUUCUAUUACAUCUCCGACCUCGUUAAAGUAAUUUCCCUAAGAUCGUGGU-UGAGUGGUAAUCGGGAAUUGG--------------------------- .(((..(((((...((.......))(((.((((((.((....((........)).....)).)))-)).).))))))))..)))...--------------------------- ( -15.40, z-score = 1.11, R) >consensus UCUUUACGAUUGCGUGGCACUUAAGAUUCCCAAUCUCACUAAGUGAGUUUUUCUAAGUGCUUGGU_UGCUUGCUAUUUGGCAAGCGGAAUCCCGGGGACUGCCACGCCCCCCC_ ...............(((((((.(((........(((((...)))))....))))))))))......(((((((....)))))))............................. (-10.01 = -11.20 + 1.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:32 2011