| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,189,323 – 18,189,418 |
| Length | 95 |
| Max. P | 0.734540 |

| Location | 18,189,323 – 18,189,418 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.15 |
| Shannon entropy | 0.32176 |
| G+C content | 0.53284 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -25.25 |
| Energy contribution | -26.66 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18189323 95 + 23011544 GCUACUGUU--UUCGAUUUCUGGUGACCAGGGCCGUAAUGUAUAAUAGAUGGCCCCGG---AUCGGAUGCGAAUUGAGGCAGUGGCCGUGGAGUA-CCCUC-------------- (((((((((--((((...((((((..((.(((((((............))))))).))---))))))..))))....)))))))))...((....-.))..-------------- ( -35.40, z-score = -2.16, R) >droSim1.chr2L 17868019 95 + 22036055 GCUACUGUU--UUCGAUUUCUGGUGACCAGGGCCGUAAUGUAUAAUAGAUGGCCCCGG---AUCGGAUGCGAAUUGAGGCAGUGGCCGUAGAGUA-CCCUC-------------- (((((((((--((((...((((((..((.(((((((............))))))).))---))))))..))))....)))))))))....(((..-..)))-------------- ( -35.10, z-score = -2.41, R) >droSec1.super_7 1814890 95 + 3727775 GCUACUGUU--UUCGAUUUCUGGUGACCAGGGCCGUAAUGUAUAAUAGAUGGCCCCGG---AUCGGAUGCGAAUUGAGGCAGUGGCCGUGGAGUA-CCCUC-------------- (((((((((--((((...((((((..((.(((((((............))))))).))---))))))..))))....)))))))))...((....-.))..-------------- ( -35.40, z-score = -2.16, R) >droYak2.chr2R 4695055 95 + 21139217 GCUACUGUU--UUCGAUUUCUGGUGACCAGGGCCGUAAUGUAUAAUAGAUGGCCCCGG---AUCGGAUGCGAAUUGAGGCAGUGACCGUGGAGUG-CCCUC-------------- ..(((((((--((((...((((((..((.(((((((............))))))).))---))))))..))))....))))))).....((....-.))..-------------- ( -29.50, z-score = -0.62, R) >droEre2.scaffold_4845 16508390 95 - 22589142 GCUACUGUU--UUCGAUUCCUGGUGACCAGGGCCGUAAUGUAUAAUAGAUGGCCCCGG---AUCGGAUGCGAAUUGAGGCAGUGACCGUGGAGUA-CCCUC-------------- ..(((((((--(.(((((((...(((((.(((((((............))))))).))---.)))...).)))))))))))))).....((....-.))..-------------- ( -30.10, z-score = -0.89, R) >droAna3.scaffold_12916 2118085 99 + 16180835 GCUACUCUGGGUUCGAUUUCUGGUGACCAGGGCCGUAAUGUAUAAUAGAUGGCCCCUGU--AUCGGAUGCGAAUGGAGGCAAUGCUCACUCAGUAGCCCUC-------------- ((((((..(.(((((...(((((((....(((((((............)))))))...)--))))))..))))).(((......))).)..))))))....-------------- ( -34.10, z-score = -1.32, R) >droPer1.super_1 4140502 113 - 10282868 --CGUAGCGGACCGGAGUUCUGGUGACCAGGGCCGUAAUGUAUAAUAGAUGGCCCCAAUGUAUCGGAUGCGAAUUGAGGCAAUACUCAUAGCACACAGAGAGCGGCUCCACUCCU --......(((..((((((((((((....(((((((............))))))).....)))))))(((....((((......))))..)))...........)))))..))). ( -35.90, z-score = -0.84, R) >consensus GCUACUGUU__UUCGAUUUCUGGUGACCAGGGCCGUAAUGUAUAAUAGAUGGCCCCGG___AUCGGAUGCGAAUUGAGGCAGUGGCCGUGGAGUA_CCCUC______________ ..(((((((..((((...((((((..((.(((((((............))))))).))...))))))..))))....)))))))............................... (-25.25 = -26.66 + 1.41)

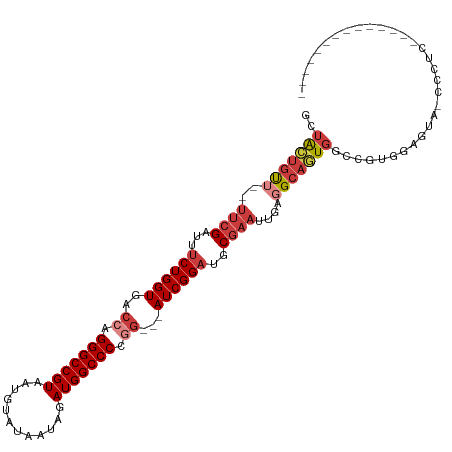

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:31 2011