
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,161,111 – 18,161,209 |
| Length | 98 |
| Max. P | 0.548397 |

| Location | 18,161,111 – 18,161,209 |
|---|---|
| Length | 98 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 65.85 |
| Shannon entropy | 0.71781 |
| G+C content | 0.37837 |
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -8.06 |
| Energy contribution | -7.97 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18161111 98 + 23011544 UCUUCCUUGGCAGUCUGUAGAUUGUCAU-AAAAUGAAAAG-----AAAUUCGAUUCGAUUAC-CAAAGGGAAAGAUCGA-GGAAGUUACUUCCCAAGCCAAAUAAU .(((((.((((((((....)))))))).-...........-----.........((((((.(-(....))...))))))-)))))..................... ( -21.00, z-score = -1.23, R) >droMoj3.scaffold_6500 26703240 86 + 32352404 CUUCUCUUGGCAGUCUGUAGAUUGUCUCAUAACUUAACAACAACAAAAUUCGAAAGGUUUAGGUUUAGUUAGAACUUAGCAAUAAC-------------------- ........(((((((....)))))))((.(((((.(((...(((......(....))))...))).))))))).............-------------------- ( -13.40, z-score = -0.12, R) >droVir3.scaffold_12963 17996309 77 + 20206255 CUUUCCUUGGCAGUCUGUAGAUUGUCACAAAACUCAACAA-----AAAUUCGAGAGGUUCAGAU------AGAACUUAGCAGAAGAAC------------------ .((((..((((((((....)))))))).............-----......(..((((((....------.))))))..).))))...------------------ ( -16.00, z-score = -0.76, R) >droPer1.super_1 4110397 101 - 10282868 UUUUUUUUGGCAGUCUGUAGAUUGUCACCAAAAUCAAAAG-----AAAUUCGCUUCGUUUACACAGGGAGGGGGAAAGAGGAAAAAUAUUUCUUCUGCCUGAUAUU .......((((((((....)))))))).....((((....-----...(((.((((.((.....)).)))).))).((((((((....))))))))...))))... ( -23.70, z-score = -1.01, R) >dp4.chr4_group3 7005261 101 - 11692001 UUUUUUUUGGCAGUCUGUAGAUUGUCACCAAAAUCAAAAG-----AAAUUCGCUUCGUUUACACAGGGAGGGGGAAAGAGGAAAAAUAUUUCUUCUGCCUGAUAUU .......((((((((....)))))))).....((((....-----...(((.((((.((.....)).)))).))).((((((((....))))))))...))))... ( -23.70, z-score = -1.01, R) >droAna3.scaffold_12916 2092698 92 + 16180835 UCUUGUUUGGCAGUUAGUAGAUUGUCACCAAAAUGAAAAG-----AAAUUCGUUUAUAGUAC-CAGGAGAGGGGGU--------GCCUUUUUGGCGGCCAAAUAAU (((((..((((((((....))))))))...(((((((...-----...))))))).......-)))))....((.(--------(((.....)))).))....... ( -24.20, z-score = -1.55, R) >droEre2.scaffold_4845 16479787 94 - 22589142 UUUUGCUUGGCAGUCUGUAGAUUGUCAC-AAAAUGAAAAG-----AAAUUCGAUUCGAAUAC-CACGGCGAAAGA-----GGAAGUUGUUUCCCUAGCCAAAUAAU (((((..((((((((....)))))))))-)))).......-----..(((((...)))))..-...(((...((.-----(((((...))))))).)))....... ( -20.90, z-score = -1.31, R) >droYak2.chr2R 4666666 93 + 21139217 UUUUCCUUGGCAGUCUGUAGAUUGUCUC-AAAAUGAAAAG-----AAAUUCGAUUAGAUUAC-CAGGGCGAAUGG------GAAGAUAUUUCCGUAGCCAAAUAAU ......((((.((((((..((...(((.-.........))-----)...))...)))))).)-)))(((..((((------(((....))))))).)))....... ( -22.50, z-score = -1.69, R) >droSec1.super_7 1786595 98 + 3727775 UCUUCCUUGGCAGUCUGUAGAUUGUCAC-AAAAUGAAAAG-----GAAUUCGAUUCGAUUAC-CAGGGGGAAAGAUCGA-GGAAGUUACUUCCCAAGCCAAAUAAU (((((((((((((((....)))))))).-..........(-----(...(((...)))...)-))))))))......(.-(((((...))))))............ ( -23.20, z-score = -0.56, R) >droSim1.chr2L 17839045 98 + 22036055 UCUUCCUUGGCAGUCUGUAGAUUGUCAC-AAAAUGAAAAG-----AAAUUCGAUUCGAUUAC-CAGGGGGAAAGAUCGA-GGAAGUUACUUCCCAAGCCAAAUAAU (((((((((((((((....)))))))).-....((((..(-----.....)..)))).....-.)))))))......(.-(((((...))))))............ ( -21.70, z-score = -0.50, R) >consensus UUUUCCUUGGCAGUCUGUAGAUUGUCAC_AAAAUGAAAAG_____AAAUUCGAUUCGAUUAC_CAGGGGGAAAGAU_GA_GGAAGAUAUUUCCCAAGCCAAAUAAU ........(((((((....)))))))................................................................................ ( -8.06 = -7.97 + -0.09)

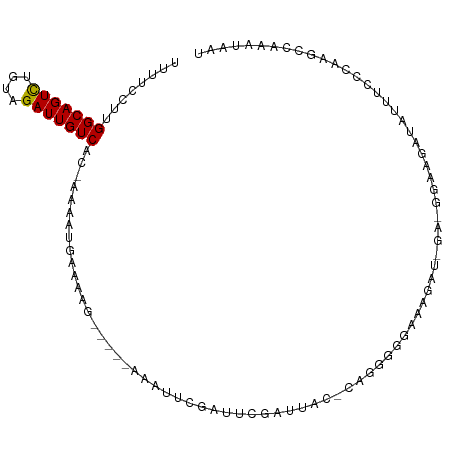
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:27 2011