| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,160,584 – 18,160,689 |
| Length | 105 |
| Max. P | 0.571616 |

| Location | 18,160,584 – 18,160,689 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.24 |
| Shannon entropy | 0.59695 |
| G+C content | 0.38159 |
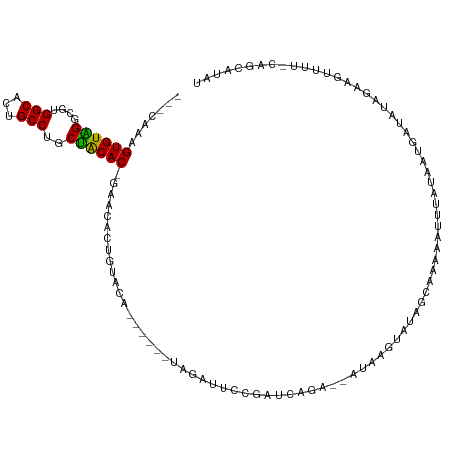
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -11.21 |
| Energy contribution | -10.47 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
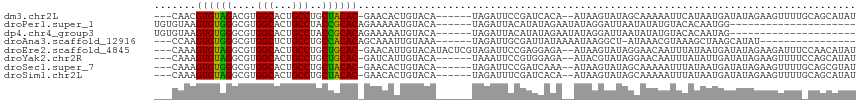
>dm3.chr2L 18160584 105 - 23011544 ---CAACGUGUAGACGUGGCACUGCCUGCUACAC-GAACACUGUACA------UAGAUUCCGAUCACA--AUAAGUAUAGCAAAAAUUCAUAAUGAUAUAGAAGUUUUGCAGCAUAU ---...(((((((.((.(((...)))))))))))-).....(((...------..(((....)))...--.........((((((.((((((....))).))).)))))).)))... ( -22.30, z-score = -1.01, R) >droPer1.super_1 4109799 90 + 10282868 UGUGUAAGUGUGGGCGUGGCACUGCCUACCGCACAGAAAAAUGUACA------UAGAUUACAUAUAGAAUAUAGGAUUAAUAUAUGUACACAAUGG--------------------- (((((..(.(((((((......)))))))))))))......((((((------((.((((..((((...))))....)))).))))))))......--------------------- ( -22.10, z-score = -1.12, R) >dp4.chr4_group3 7004680 90 + 11692001 UGUGUAAGUGUGGGCGUGGCACUGCCUACCGCACAGAAAAAUGUACA------UAGAUUACAUAUAGAAUAUAGGAUUAAUAUAUGUACACAAUAG--------------------- (((((..(.(((((((......)))))))))))))......((((((------((.((((..((((...))))....)))).))))))))......--------------------- ( -22.10, z-score = -1.41, R) >droAna3.scaffold_12916 2092156 91 - 16180835 ---CCAAGUGUGGGCGUGGCUCUGCCUGCCACACAGCAAAUUGUAAA------UAGAUUGCGAUUAUAAAAUAAGGCU-AUAAACGUAAAGCUAAGCAUAU---------------- ---....((((((.((.(((...))))))))))).((.((((((((.------....)))))))).........((((-.((....)).))))..))....---------------- ( -23.40, z-score = -1.06, R) >droEre2.scaffold_4845 16479254 111 + 22589142 ---CAAAGUGUAGGCGUGGCACUGCCUGCUGCAC-GAACAUUGUACAUACUCGUAGAUUCCGAGGAGA--AUAAGUAUAGGAACAAUUUAUAAUGAUAUAGAAGAUUUCCAACAUAU ---....((((((.((.(((...)))))))))))-......(((((...((((.......))))....--....)))))((((...((((((....))))))....))))....... ( -21.20, z-score = -0.10, R) >droYak2.chr2R 4666139 105 - 21139217 ---CAAAGUGUAGGCGUGGCACUGCCUGCUGCAC-GAUCAUUGUACA------UAAAUUCCGUGGAGA--AUACGUAUAGGAACAAUUUAUAUUGAUAUAGAAGUUUUCCAGCAUAU ---......(((((((......)))))))(((..-......(((((.------((..(((....))).--.)).)))))((((.(((((.(((....))).))))))))).)))... ( -22.20, z-score = -0.03, R) >droSec1.super_7 1786068 105 - 3727775 ---CAAAGUGUGGGCGUGGCACUGCCUGCUACAC-GAACACUGUACA------UAGAUUCCGAUCAAA--AUAAGUAUAGCAAAAAUUUAUAAUGAUAUAGAAGUUUUGCAGCGUAU ---...(((((..(.((((((.....)))))).)-..)))))((((.------..(((....)))...--.........((((((.((((((....))))))..))))))...)))) ( -23.80, z-score = -0.89, R) >droSim1.chr2L 17838518 105 - 22036055 ---CAAAGUGUAGGCGUGGCACUGCCUGCUACAC-GAACACUGUACA------UAGAUUUCGAUCACA--AUAAGUAUAGCAAAAAUUUAUAAUGAUAUAGAAGUUUUGCAGCAUAU ---....((((((.((.(((...)))))))))))-.....(((((..------.(((((((.((((..--((((((.........))))))..))))...))))))))))))..... ( -25.00, z-score = -1.66, R) >consensus ___CAAAGUGUAGGCGUGGCACUGCCUGCUACAC_GAACACUGUACA______UAGAUUCCGAUCAGA__AUAAGUAUAGCAAAAAUUUAUAAUGAUAUAGAAGUUUU_CAGCAUAU .......((((((....(((...)))..))))))................................................................................... (-11.21 = -10.47 + -0.73)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:26 2011