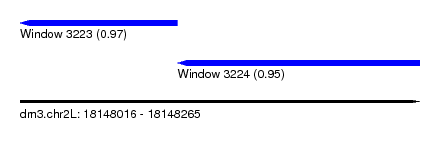
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,148,016 – 18,148,265 |
| Length | 249 |
| Max. P | 0.969323 |

| Location | 18,148,016 – 18,148,114 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 93.49 |
| Shannon entropy | 0.09022 |
| G+C content | 0.46497 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -26.27 |
| Energy contribution | -26.93 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.969323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18148016 98 - 23011544 GACUCAAAUGUAUCUGGGGAAACCAUCUUUUUUGGCCAUUUUGGCCAAAGGCGAAAAAGGGGAA-AGAACUAGAUACAUUCGGGCCUAACCCGGACCUG ........((((((((((....)).((.(((((((((.....))))))))).))..........-.....))))))))((((((.....)))))).... ( -33.90, z-score = -2.91, R) >droSec1.super_7 1773770 94 - 3727775 GACUCGAAUGUAUCUGGGGAAACCAUCUUUUU-----AUUUUGGCCAAAGGCGAAAAAGGGGAAAAGAGCUAGAUACAUUCGGGCCUAACCCGGACCUG ..((((((((((((((((....)).(((((((-----.((((.(((...))).))))....)))))))..))))))))))))))((......))..... ( -34.90, z-score = -3.80, R) >droSim1.chr2L 17824177 94 - 22036055 GACUCGAAUGUAUCUGGGGAAACCAUCUUUUU-----AUUUUGGCCAAAGGAGAAAAAGGGGAACAGAACUAGAUACAUUCGGGCCUAACCCGGACCUG ..((((((((((((((((....)).(((((((-----.((((......)))).)))))))..........))))))))))))))((......))..... ( -29.20, z-score = -2.30, R) >consensus GACUCGAAUGUAUCUGGGGAAACCAUCUUUUU_____AUUUUGGCCAAAGGCGAAAAAGGGGAA_AGAACUAGAUACAUUCGGGCCUAACCCGGACCUG ..((((((((((((((((....)).(((.(((......((((.(((...))).))))....))).)))..))))))))))))))((......))..... (-26.27 = -26.93 + 0.67)

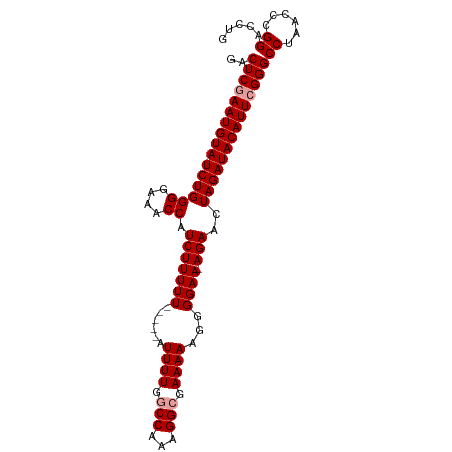
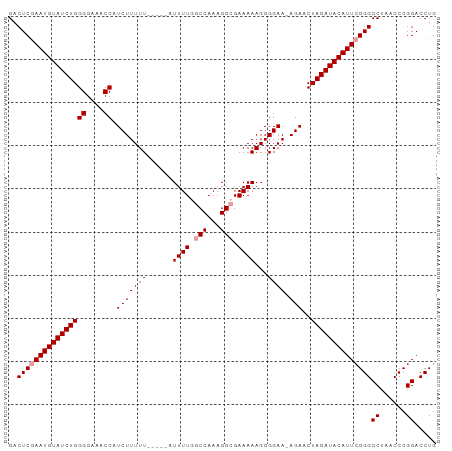
| Location | 18,148,114 – 18,148,265 |
|---|---|
| Length | 151 |
| Sequences | 6 |
| Columns | 157 |
| Reading direction | reverse |
| Mean pairwise identity | 71.37 |
| Shannon entropy | 0.52467 |
| G+C content | 0.44030 |
| Mean single sequence MFE | -36.51 |
| Consensus MFE | -19.66 |
| Energy contribution | -19.55 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
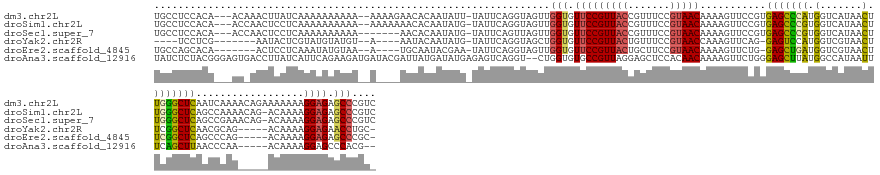
>dm3.chr2L 18148114 151 - 23011544 UGCCUCCACA---ACAAACUUAUCAAAAAAAAAA--AAAAGAACACAAUAUU-UAUUCAGGUAGUUGGUGUUCCGUUACCGUUUCCGUAACAAAAGUUCCGUGAGCCCAUGGUCAUAACUUGGGCUCAAUCAAAACAGAAAAAAAGGAGAGCCCGUC .((((((...---...(((((.............--....((((((..((((-(....)))))....)))))).(((((.......)))))..)))))...((((((((.(((....))))))))))).................)))).))..... ( -37.00, z-score = -3.35, R) >droSim1.chr2L 17824271 150 - 22036055 UGCCUCCACA---ACCAACUCCUCAAAAAAAAAA--AAAAAAACACAAUAUG-UAUUCAGGUAGUUGGUGUUCCGUUACCGUUUCCGUAACAAAAGUUCCGUGAGCCCGUGGUCAUAACUUGGGCUCAGCCAAAACAG-ACAAAAGGAGAGCCCGUC .((((((...---((((((((((...........--................-.....))).)))))))(((..(((((.......)))))..........((((((((.(((....))))))))))).........)-))....)))).))..... ( -36.65, z-score = -2.35, R) >droSec1.super_7 1773864 145 - 3727775 UGCCUCCACA---ACCAACUCCUCAAAAAAAAAA-------AACACAAUAUG-UAUUCAGUUAGUUGGUGUUCCGUUACCGUUUCCGUAACAAAAGUUCCGUGAGCCCGUGGUCAUAACUUGGGCUCAGCCGAAACAG-ACAAAAGGAGAGCCCGUC .((((((...---(((((((..............-------...........-.........)))))))(((..(((((.......)))))....(((.((((((((((.(((....)))))))))))..)).))).)-))....)))).))..... ( -34.98, z-score = -1.80, R) >droYak2.chr2R 4652860 132 - 21139217 ----UCCUCG-------AAUACUCGUAUGUAUGU--A----AAUACAAUAUG-UAUUCAGGUAGCUGGUGUUCCGUUACUGUUUCCGUAACCAAAGUUCAG-GAGUCCAUGGUCGUAACUUCGGCUCAACGCAG-----ACAAAAGGAGAACCUGC- ----.(((.(-------(((((..((.(((((..--.----.)))))))..)-))))))))..((.(((.((((.....(((((.(((..((........)-)......((((((......)))).))))).))-----)))...)))).))).))- ( -33.60, z-score = -1.16, R) >droEre2.scaffold_4845 16467408 136 + 22589142 UGCCAGCACA-------ACUCCUCAAAUAUGUAA--A----UGCAAUACGAA-UAUUCAGGUAGUUGGUGUUCCGUUACUGCUUCCGUAACAAAAGUUCUG-GAGCUGAUGGUCGUAACUUCGGCUCAGCCCAG-----ACAAAAGGAGAGCCCGC- .((..((.((-------((((((..((((((((.--.----.....)))..)-)))).))).)))))...(((((((((.......)))))....((.(((-(.(((((..((((......)))))))))))))-----))....)))).))..))- ( -38.20, z-score = -1.48, R) >droAna3.scaffold_12916 13410196 148 + 16180835 UAUCUCUACGGGAGUGACCUUAUCAUUCAGAAGAUGAUACGAUUAUGAUAUGAGAGUCAGGU--CUGGUGUGCCGUUAGGAGCUCCACAACAAAAGUUCUGGGAGCUUAUGGCCAUAAUUUCAGCUUAACCCAA-----ACAAAAGGAGCCCACG-- .........((((((((((.(((((((.....)))))))......((((......)))))))--)..(((.(((((...(((((((..(((....)))...))))))))))))))).......)))....((..-----......))..)))...-- ( -38.60, z-score = -0.49, R) >consensus UGCCUCCACA___AC_AACUCCUCAAAAAAAAAA__A____AACACAAUAUG_UAUUCAGGUAGUUGGUGUUCCGUUACCGUUUCCGUAACAAAAGUUCCGUGAGCCCAUGGUCAUAACUUCGGCUCAACCCAA_____ACAAAAGGAGAGCCCGU_ ..................................................................(((.(((((((((.......)))))...........(((((((.(.......).)))))))..................)))).))).... (-19.66 = -19.55 + -0.11)
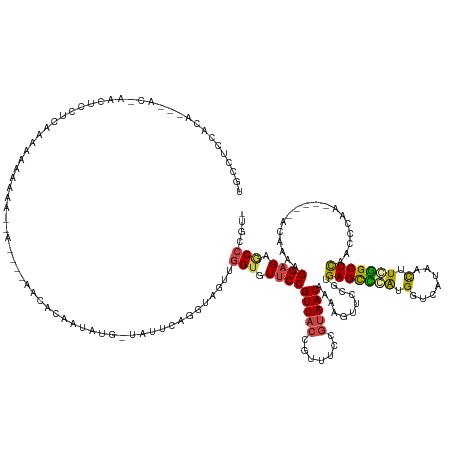
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:25 2011