| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,126,727 – 18,126,846 |
| Length | 119 |
| Max. P | 0.956198 |

| Location | 18,126,727 – 18,126,846 |
|---|---|
| Length | 119 |
| Sequences | 13 |
| Columns | 147 |
| Reading direction | forward |
| Mean pairwise identity | 84.22 |
| Shannon entropy | 0.31221 |
| G+C content | 0.42553 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.16 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
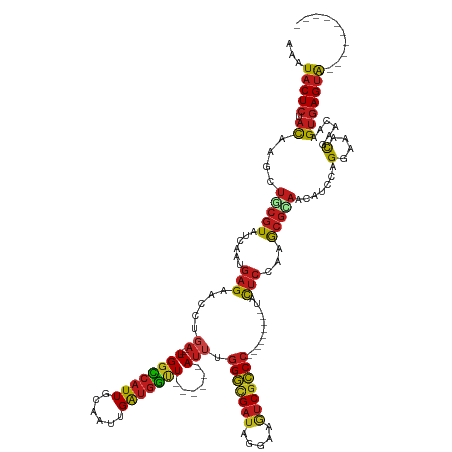
>dm3.chr2L 18126727 119 + 23011544 AAAUACUCCACAAGCUGCGUAUCAAUGAGAACCUGA---UGGCCAUUGCAAUUGAUGGUU------AUUUGGGCGAUAGGAAGUCGCCC---------UACUCCAAGCGCAAUAUCCAGGAAACGAACAAGUGAGUA---------- ...(((((.((....(((((......(((.....((---((((((((......)))))))------))).(((((((.....)))))))---------..)))...))))).......(....)......)))))))---------- ( -36.60, z-score = -2.56, R) >droSim1.chr2L 17802898 119 + 22036055 AAAUACUCCACAAGCUGCGUAUCAAUGAGAACCUGA---UGGCCAUUGCAAUUGAUGGUU------AUUUGGGCGAUAGGAAGUCGCCC---------UACUCCAAGCGCAAUAUCCAGGAAACGAACAAGUGAGUA---------- ...(((((.((....(((((......(((.....((---((((((((......)))))))------))).(((((((.....)))))))---------..)))...))))).......(....)......)))))))---------- ( -36.60, z-score = -2.56, R) >droSec1.super_7 1752424 119 + 3727775 AAAUACUCCACAAGCUGCGUAUCAAUGAGAACCUUA---UGGCCAUUGCAAUUGAUGGUU------AUUUGGGCGAUAGGAAGUCGCCC---------UACUCCAAGCGCAAUAUCCAGGAAACGAACAAGUGAGUA---------- ...(((((.((....(((((......(((......(---((((((((......)))))))------))..(((((((.....)))))))---------..)))...))))).......(....)......)))))))---------- ( -35.30, z-score = -2.37, R) >droYak2.chr2R 4631264 119 + 21139217 AAAUACUCCACAAGCUGCGUAUCAAUGAGAACCUGA---UGGCCAUUGCAAUUGAUGGUU------AUUUGGGCGAUAGGAAGUCGCCC---------UACUCCAAGCGCAAUAUCCAGGAAACGAACAAGUGAGUA---------- ...(((((.((....(((((......(((.....((---((((((((......)))))))------))).(((((((.....)))))))---------..)))...))))).......(....)......)))))))---------- ( -36.60, z-score = -2.56, R) >droEre2.scaffold_4845 16446177 119 - 22589142 AAAUACUCCACAAGCUGCGUAUCAAUGAGAACCUGA---UGGCCAUUGCAAUUGAUGGUU------AUUUGGGCGAUAGGAAGUCGCCC---------UACUCCAAGCGCAAUAUCCAGGAAACGAACAAGUGAGUA---------- ...(((((.((....(((((......(((.....((---((((((((......)))))))------))).(((((((.....)))))))---------..)))...))))).......(....)......)))))))---------- ( -36.60, z-score = -2.56, R) >droAna3.scaffold_12916 13388937 129 - 16180835 AAAUACUCUAUAAGCUACGUAUCAAUGAGAACCUGA---UGGCCAUUGCAAUCGAUGGUU------AUUUGGGCGAUAGGAAGUCGCCC---------UACUCCAAGCGGAACAUACAGGAAACGAACAAGUGAGUACUAAGGACUA ...(((((......((.(((....)))))..(((((---(((((((((....))))))))------))..(((((((.....)))))))---------...(((....))).....))))............))))).......... ( -33.80, z-score = -1.69, R) >dp4.chr4_group3 1432984 119 - 11692001 AAAUACUCUACAAGCUGCGUAUCAAUGAGAAUUUGA---UGGCCAUUGGAAUUGAUGGUU------AUUUGGGCGAUAGGAAGUCGCCC---------UACUCCAAGCGGAACAUACAGGAAACGAACAAGUGAGUA---------- ...(((((.((...((((........(((.....((---((((((((......)))))))------))).(((((((.....)))))))---------..)))...))))........(....)......)))))))---------- ( -34.60, z-score = -2.52, R) >droPer1.super_8 2497154 119 - 3966273 AAAUACUCUACAAGCUGCGUAUCAAUGAGAAUUUGA---UGGCCAUUGGAAUUGAUGGUU------AUUUGGGCGAUAGGAAGUCGCCC---------UACUCCAAGCGGAACAUACAGGAAACGAACAAGUGAGUA---------- ...(((((.((...((((........(((.....((---((((((((......)))))))------))).(((((((.....)))))))---------..)))...))))........(....)......)))))))---------- ( -34.60, z-score = -2.52, R) >droWil1.scaffold_180764 3724668 119 - 3949147 AAAUACUCUAUAAGCUACGUAUCAAUGAGAAUUUGA---UGGCCAUUGCAAUUGAUGGUU------AUUUGGGUGAUAGAAAAUCGCCA---------UAUUCGAAGCGUAACAUCCAGGAAACGAAUAAGUGAGUA---------- ...(((((.....(.(((((.((...........((---((((((((......)))))))------)))..((((((.....)))))).---------.....)).))))).).....(....)........)))))---------- ( -30.00, z-score = -2.24, R) >droVir3.scaffold_12963 17218342 119 + 20206255 AAAUACUCUACAAGCUGCGUAUCAAUGAGCAAUUGA---UGGCCAUUGGAAUUGAUGGUU------AUUUGGGUGAUAGGAAAUCGCCC---------UACUCCAAGCGUAACAUACAGGAAACGAACAAGUGAGUG---------- ...(((((.((..(.(((((((((((.....)))))---)(((((((......)))))))------....(((((((.....)))))))---------........))))).).....(....)......)))))))---------- ( -32.10, z-score = -1.87, R) >droMoj3.scaffold_6500 22258951 119 + 32352404 AAAUACUCUACAAGCUGCGUAUCAAUGAGCAAUUGA---UGGUCAUUGGAAUUGAUGGCU------AUUUGGGUGAUAGAAAAUCGCCC---------UAUUCGAAGCGCAACAUACAGGAAACGAAUAAGUGAGUA---------- ...(((((.((..(.(((((.((...........((---((((((((......)))))))------))).(((((((.....)))))))---------.....)).))))).).....(....)......)))))))---------- ( -35.80, z-score = -3.43, R) >droGri2.scaffold_15252 14643891 119 + 17193109 AAAUACUCUACAAGCUACGUAUCAAUGAGCAUUUGA---UGGCCAUUGGAAUUGAUGGAU------AUUUGGGUGAUAGAAAAUCGCCC---------UACUCCAAGCGUAACAUACAGGAAAUGAGCAAGUGAGUA---------- ...(((((.((..(((((((.((((((.((......---..))))))))......((((.------....(((((((.....)))))))---------...)))).))))..(((.......))))))..)))))))---------- ( -32.30, z-score = -2.34, R) >anoGam1.chr3R 1210748 137 - 53272125 AAGUACUCACCAAGCGCAAUAUUACCGAGAAUCUUAGCCUGGUGUUUAUUGUCGGCAAUUUGACAGAGUCGGAUGUUCACAGCUCCUCCAUAAAUAAGUUUAAUAAACGAAACAUCCAGGAAACAAACCGGUAAGCA---------- ...............((....((((((.(........((((((((((.((((..(((.((((((...)))))))))..))))...............(((.....))).))))).))))).......))))))))).---------- ( -27.76, z-score = -0.43, R) >consensus AAAUACUCUACAAGCUGCGUAUCAAUGAGAACCUGA___UGGCCAUUGCAAUUGAUGGUU______AUUUGGGCGAUAGGAAGUCGCCC_________UACUCCAAGCGCAACAUCCAGGAAACGAACAAGUGAGUA__________ ...(((((.((....(((((......(((...........(((((((......)))))))..........(((((((.....)))))))...........)))...))))).......(....)......))))))).......... (-24.26 = -24.16 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:22 2011