
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,108,628 – 18,108,720 |
| Length | 92 |
| Max. P | 0.524365 |

| Location | 18,108,628 – 18,108,720 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.20 |
| Shannon entropy | 0.38675 |
| G+C content | 0.30549 |
| Mean single sequence MFE | -15.86 |
| Consensus MFE | -11.50 |
| Energy contribution | -11.52 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
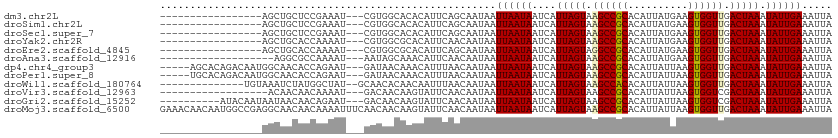
>dm3.chr2L 18108628 92 + 23011544 -----------------AGCUGCUCCGAAAU---CGUGGCACACAUUCAGCAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUGAAGUGGUUGACUAAAUAUUGAAAUUA -----------------.(.(((..((....---))..))))..............((((((....(((((.((((((..........)))))).))))).))))))..... ( -17.90, z-score = -0.79, R) >droSim1.chr2L 17784307 92 + 22036055 -----------------AGCUGCUCCGAAAU---CGUGGCACACAUUCAGCAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUGAAGUGGUUGACUAAAUAUUGAAAUUA -----------------.(.(((..((....---))..))))..............((((((....(((((.((((((..........)))))).))))).))))))..... ( -17.90, z-score = -0.79, R) >droSec1.super_7 1734580 92 + 3727775 -----------------AGCUGCUCCGAAAU---CGUGGCACACAUUCAGCAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUGAAGUGGUUGACUAAAUAUUGAAAUUA -----------------.(.(((..((....---))..))))..............((((((....(((((.((((((..........)))))).))))).))))))..... ( -17.90, z-score = -0.79, R) >droYak2.chr2R 18157301 92 - 21139217 -----------------AGCUGCACCAAAAU---CGUGGCGCACAUUCAACAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUGAAGUGGUUGACUAAAUAUUGAAAUUA -----------------.(.(((.(((....---..))).))))............((((((....(((((.((((((..........)))))).))))).))))))..... ( -18.10, z-score = -1.12, R) >droEre2.scaffold_4845 16425966 92 - 22589142 -----------------AGCUGCACCAAAAU---CGUGGCGCACAUUCAGCAAUAAUUAAUAAUCAUUAGUAGGCCGCACAUUAUGAAGUGGUUGACUAAAUAUUGAAAUUA -----------------.(((((.(((....---..))).))......))).....((((((....(((((..(((((..........)))))..))))).))))))..... ( -18.60, z-score = -0.54, R) >droAna3.scaffold_12916 13372206 90 - 16180835 -------------------AGGCGCCAAAAU---AAUAGCAAACAUUCAACAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUGAAGUGGUUGACUAAAUAUUGAAAUUA -------------------..((........---....))................((((((....(((((.((((((..........)))))).))))).))))))..... ( -12.20, z-score = -0.17, R) >dp4.chr4_group3 1410444 104 - 11692001 -----AGCACAGACAAUGGCAACACCAGAAU---GAUAACAAACAUUUAACAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUUAAGUGGUUGACUAAAUAUUGAAAUUA -----.((.((.....)))).......((((---(........)))))........((((((....(((((.((((((..........)))))).))))).))))))..... ( -15.40, z-score = -1.34, R) >droPer1.super_8 2474576 104 - 3966273 -----UGCACAGACAAUGGCAACACCAGAAU---GAUAACAAACAUUUAACAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUUAAGUGGUUGACUAAAUAUUGAAAUUA -----(((.((.....)))))......((((---(........)))))........((((((....(((((.((((((..........)))))).))))).))))))..... ( -15.90, z-score = -1.32, R) >droWil1.scaffold_180764 3684854 96 - 3949147 --------------UGUAAAUCUAUGGCUAU--GCAACACAACAAUUUAACAAUAAUUAAUAAUCAUUAGUAAGCCACACAUUAUUAAGUGGUUGACUAAAUAUUGAAAUUA --------------.................--.......................((((((....(((((.((((((..........)))))).))))).))))))..... ( -12.00, z-score = 0.12, R) >droVir3.scaffold_12963 17195808 91 + 20206255 ------------------ACAACAACAAAAU---GACAACAAGUAUUCAACAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUUAAGUGGUCGACUAAAUAUUGAAAUUA ------------------.............---......................((((((....(((((..(((((..........)))))..))))).))))))..... ( -11.40, z-score = -1.38, R) >droGri2.scaffold_15252 14623051 99 + 17193109 ----------AUACAAUAAUAACAACAGAAU---GACAACAAGUAUUCAACAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUUAAGUGGUCGACUAAAUAUUGAAAUUA ----------...(((((.........((((---(........)))))..................(((((..(((((..........)))))..))))).)))))...... ( -15.50, z-score = -2.60, R) >droMoj3.scaffold_6500 22232615 112 + 32352404 GAAACAACAAUGGCCGAGGCAACAACAAAAUUUCAACAACAAGUAUUCAACAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUUAAGUGGUUGACUAAAUAUUGAAAUUA ............((....))........((((((((......((.....))...............(((((.((((((..........)))))).)))))...)))))))). ( -17.50, z-score = -1.86, R) >consensus _________________AGCUGCACCAAAAU___CAUAACAAACAUUCAACAAUAAUUAAUAAUCAUUAGUAAGCCGCACAUUAUGAAGUGGUUGACUAAAUAUUGAAAUUA ........................................................((((((....(((((.((((((..........)))))).))))).))))))..... (-11.50 = -11.52 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:19 2011