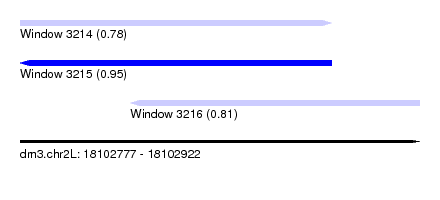
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,102,777 – 18,102,922 |
| Length | 145 |
| Max. P | 0.947149 |

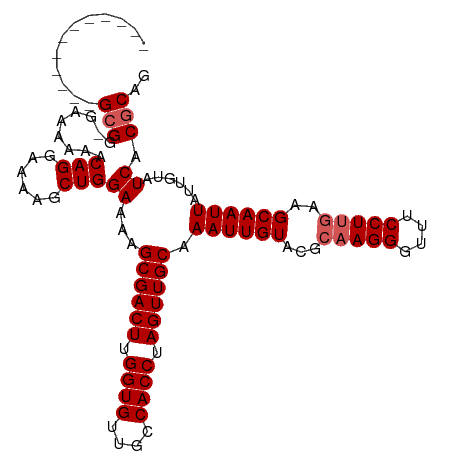
| Location | 18,102,777 – 18,102,890 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 90.71 |
| Shannon entropy | 0.15424 |
| G+C content | 0.47151 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -27.26 |
| Energy contribution | -27.86 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
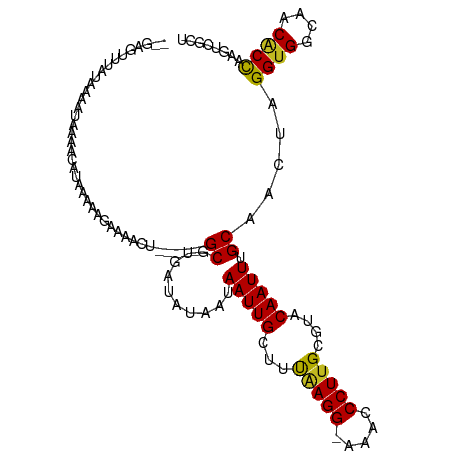
>dm3.chr2L 18102777 113 + 23011544 GUGGGCGGGGGGGGGGGGGGUGAACAGGAAAAGCUGGAAAAGCGACUUGGUGUUGCCACCUAGUUGCAAAUUGUACGCAAGGGUUUCCUUAAAGCAAUUAUUGUAUCACGCAG .................(.((((((((.....((((((((.((((((.((((....)))).))))))........(....)..)))))....))).....)))).)))).).. ( -30.40, z-score = 0.24, R) >droEre2.scaffold_4845 16420147 100 - 22589142 ------------GCGG-GGAAAAACAGGAAAAGCUGGAAAAGCGACUUGGUGUUGCCACCUAGUUGCAAAUUGUACGCAAGGGUUUCCUCGCAGCAAUUAUUGUAUCACGCAG ------------((((-(((((..(((......))).....((((((.((((....)))).))))))........(....)..)))))))))..................... ( -31.70, z-score = -1.81, R) >droYak2.chr2R 18151382 100 - 21139217 ------------GCGG-GAAAAAACAGGAAAAGCUGGAAAAGCGACUUGGUGUUGCCACCUAGUUGCAAAUUGUACGCAAGGGUUUCCUUGAAGCAAUUAUUGUAUCACGCAG ------------(((.-((.....(((......))).....((((((.((((....)))).)))))).((((((...(((((....)))))..))))))......)).))).. ( -31.40, z-score = -2.30, R) >droSec1.super_7 1728861 100 + 3727775 ------------GCGG-AAAAAAACAGGAAAAGCUGGAAAAGCGACUUGGUGUUGCCACCUAGUUGCAAAUUGUACGCAAGGGUUUCCUUGGAGCAAUUAUUGUAUCACGCAG ------------((((-(......(((......))).....((((((.((((....)))).)))))).((((((.(.(((((....)))))).))))))......)).))).. ( -31.60, z-score = -2.36, R) >droSim1.chr2L 17778501 101 + 22036055 ------------GCGGGAAAAAAACAGGAAAAGCUGGAAAAGCGACUUGGUGUUGCCACCUAGUUGCAAAUUGUACGCAAGGGUUUCCUUGGAGCAAUUAUUGUAUCACGCAG ------------(((.((......(((......))).....((((((.((((....)))).)))))).((((((.(.(((((....)))))).))))))......)).))).. ( -31.70, z-score = -2.17, R) >consensus ____________GCGG_GAAAAAACAGGAAAAGCUGGAAAAGCGACUUGGUGUUGCCACCUAGUUGCAAAUUGUACGCAAGGGUUUCCUUGAAGCAAUUAUUGUAUCACGCAG ............(((.........(((......)))((...((((((.((((....)))).)))))).((((((...(((((....)))))..))))))......)).))).. (-27.26 = -27.86 + 0.60)

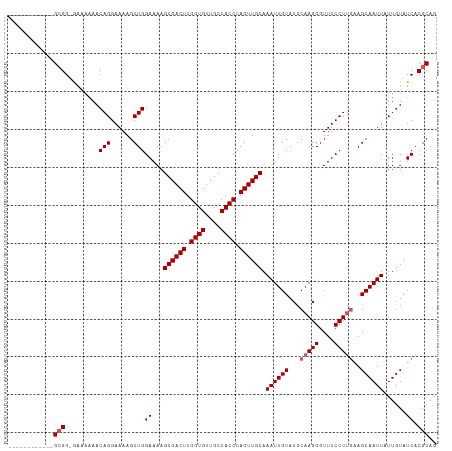
| Location | 18,102,777 – 18,102,890 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.71 |
| Shannon entropy | 0.15424 |
| G+C content | 0.47151 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -22.38 |
| Energy contribution | -22.22 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.947149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18102777 113 - 23011544 CUGCGUGAUACAAUAAUUGCUUUAAGGAAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCUUUUCCAGCUUUUCCUGUUCACCCCCCCCCCCCCCGCCCAC ....((((.(((..((..(((.(((((....)))))..........((.(((.((((....)))).))).))......)))..))..)))))))................... ( -25.50, z-score = -2.32, R) >droEre2.scaffold_4845 16420147 100 + 22589142 CUGCGUGAUACAAUAAUUGCUGCGAGGAAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCUUUUCCAGCUUUUCCUGUUUUUCC-CCGC------------ ..(((.((.(((..((..(((((((((....)))))).........((.(((.((((....)))).))).))......)))..))..)))...)).-.)))------------ ( -30.20, z-score = -2.85, R) >droYak2.chr2R 18151382 100 + 21139217 CUGCGUGAUACAAUAAUUGCUUCAAGGAAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCUUUUCCAGCUUUUCCUGUUUUUUC-CCGC------------ ..(((.((.(((..((..(((.(((((....)))))..........((.(((.((((....)))).))).))......)))..))..)))....))-.)))------------ ( -26.40, z-score = -2.33, R) >droSec1.super_7 1728861 100 - 3727775 CUGCGUGAUACAAUAAUUGCUCCAAGGAAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCUUUUCCAGCUUUUCCUGUUUUUUU-CCGC------------ ..(((.((.(((..((..(((.(((((....)))))..........((.(((.((((....)))).))).))......)))..))..))).....)-))))------------ ( -25.90, z-score = -2.16, R) >droSim1.chr2L 17778501 101 - 22036055 CUGCGUGAUACAAUAAUUGCUCCAAGGAAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCUUUUCCAGCUUUUCCUGUUUUUUUCCCGC------------ ..(((.((.(((..((..(((.(((((....)))))..........((.(((.((((....)))).))).))......)))..))..))).....)).)))------------ ( -25.90, z-score = -2.15, R) >consensus CUGCGUGAUACAAUAAUUGCUCCAAGGAAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCUUUUCCAGCUUUUCCUGUUUUUUC_CCGC____________ ..(((.((.(((..((..(((.(((((....)))))..........((.(((.((((....)))).))).))......)))..))..)))...))...)))............ (-22.38 = -22.22 + -0.16)

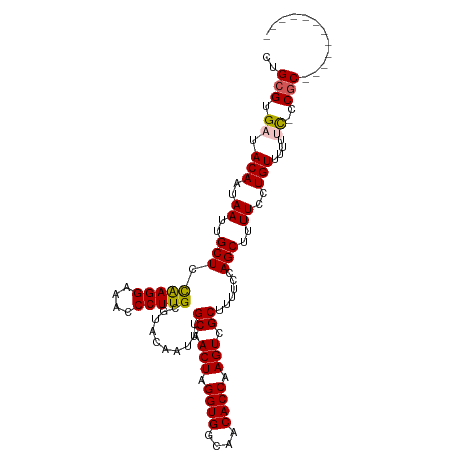
| Location | 18,102,817 – 18,102,922 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 71.55 |
| Shannon entropy | 0.59546 |
| G+C content | 0.40963 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -11.63 |
| Energy contribution | -11.12 |
| Covariance contribution | -0.51 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18102817 105 - 23011544 AGGAGUUUAUAAAAUAAAAUGUAACAACAAAACU------GCGUGAUACAAUAAUUGCUUUAAGG-AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCU .(.(((((.....................)))))------.)(..((......))..)..(((((-....)))))..........((.(((.((((....)))).))).)). ( -20.80, z-score = -0.63, R) >droPer1.super_8 2468911 111 + 3966273 GCGAGUCCAUAAAAAGCAACAACUAAAGAAAAGUGCUCAGGCUUGAUAUCAUAAUUGCUUUAAGG-AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCGCCCCACU ((((((......(((((((...........((((......))))..........)))))))((((-....)))).......)))))).....((((....))))........ ( -23.60, z-score = -0.52, R) >dp4.chr4_group3 1404807 111 + 11692001 GCGAGUCCAUAAAAAGCAACAACUAAAGAAAAGUGCUCAGGCUUGAUAUCAUAAUUGCUUUAAGG-AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCGCCCCACU ((((((......(((((((...........((((......))))..........)))))))((((-....)))).......)))))).....((((....))))........ ( -23.60, z-score = -0.52, R) >droAna3.scaffold_12916 13367156 102 + 16180835 -------CGGAGUAAAUAA-AAUAAAAAGUAACUAAAC-UGCGUGAUAUAAUAAUUGCUUUAAGG-AAACCCUCGCGUACAAUUUGCAACUAGGUGGCAACACCAGGACUCG -------..((((......-........((((......-(((((((.....(((.....))).(.-...)..)))))))....))))..((.((((....)))).)))))). ( -22.20, z-score = -1.81, R) >droEre2.scaffold_4845 16420174 105 + 22589142 AGGAGUUUAUAAAAUAAAAUGUAGCAGCAAAACU------GCGUGAUACAAUAAUUGCUGCGAGG-AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCU ...................((((((((.....))------))(..((......))..).((((((-....)))))).))))....((.(((.((((....)))).))).)). ( -31.00, z-score = -2.54, R) >droYak2.chr2R 18151409 105 + 21139217 AGGAGUUUAUAAAAUAAAAUGUAGCAGCAAAACU------GCGUGAUACAAUAAUUGCUUCAAGG-AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCU ...................((((((((.....))------))(..((......))..)..(((((-....)))))..))))....((.(((.((((....)))).))).)). ( -26.70, z-score = -1.74, R) >droSec1.super_7 1728888 105 - 3727775 AAGAGUUUAUAAAAUAAAAUGUAACAGCAAAACU------GCGUGAUACAAUAAUUGCUCCAAGG-AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCU ...................((((.(((((....)------)).)).))))..(((((..((((((-....))))).)..))))).((.(((.((((....)))).))).)). ( -27.90, z-score = -2.99, R) >droSim1.chr2L 17778529 105 - 22036055 AGGAGUUUAUAAAAUAAAAUGUAACAGCAAAACU------GCGUGAUACAAUAAUUGCUCCAAGG-AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCU ...................((((.(((((....)------)).)).))))..(((((..((((((-....))))).)..))))).((.(((.((((....)))).))).)). ( -27.90, z-score = -2.58, R) >droVir3.scaffold_12963 17189464 90 - 20206255 ------------UCGAGUGCAUAAAAAGGAAACU------GC---GUGUUGUAAUUGCUUUAAGG-AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACGCUGUCGCGCU ------------...(((((......((....))------((---(.((((((...((...((((-....)))))).)))))).))).....((((....))))...))))) ( -26.80, z-score = -1.22, R) >droMoj3.scaffold_6500 22224259 94 - 32352404 ---------UUGGCGAGUGCAUAAAAAGGAAAAU------GC---GUGUUGUAAUUGCUUUAAGGAAAACCCUUGCGUACAAUUUGCAGCUAGGUGGCAACGCUGUCGCGCU ---------......(((((.......((....(------((---(.((((((...((...((((.....)))))).)))))).)))).)).((((....))))...))))) ( -26.00, z-score = 0.06, R) >droGri2.scaffold_15252 14617112 90 - 17193109 ------------UGUAGUGCAUAAAAAGGAAACU------GC---GUAUUGUAAUUGCUUUAAGG-AAACCCUUGCGUACAAUUUGCGACUAGGUGGCAACGCUGUCGCGCA ------------....((((......((....))------((---(.((((((...((...((((-....)))))).)))))).)))(((..((((....))))))))))). ( -29.20, z-score = -1.98, R) >droWil1.scaffold_180764 282386 98 - 3949147 ---AAGUGUAUUAAAAGUAAAGAGCGGGAAUACU------GC----UGUAAUAAUUGCUUUAAGG-AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACUCUACUCUAUG ---.((.(((...((((((((.(((((.....))------))----).).....)))))))...(-..(((.(((((.......)))))...)))..).....))).))... ( -19.10, z-score = 0.18, R) >consensus __GAGUUUAUAAAAUAAAACAUAAAAAGAAAACU______GCGUGAUAUAAUAAUUGCUUUAAGG_AAACCCUUGCGUACAAUUUGCAACUAGGUGGCAACACCAAGUCGCU ....................................................(((((...(((((.....)))))....)))))........((((....))))........ (-11.63 = -11.12 + -0.51)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:18 2011