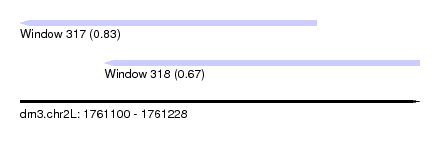
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,761,100 – 1,761,228 |
| Length | 128 |
| Max. P | 0.829028 |

| Location | 1,761,100 – 1,761,195 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.18 |
| Shannon entropy | 0.25450 |
| G+C content | 0.34990 |
| Mean single sequence MFE | -20.80 |
| Consensus MFE | -12.17 |
| Energy contribution | -13.36 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
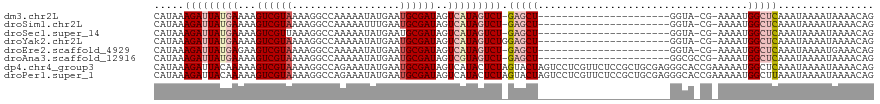
>dm3.chr2L 1761100 95 - 23011544 CAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCU-GAGCU----------------------GGUA-CG-AAAAUGGCUCAAAUAAAAUAAAACAG .....(((((((((...((((((..................))))))..)))))))))-(((((----------------------(...-..-....))))))................ ( -20.07, z-score = -3.43, R) >droSim1.chr2L 1661269 95 - 22036055 CAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUUUGAAUGCGAUAGUCAUAGUCU-GAGCU----------------------GGUA-CG-AAAAUGGCUCAAAUAAAAUAAAACAG .....(((((((((...((((((.(((........)))...))))))..)))))))))-(((((----------------------(...-..-....))))))................ ( -20.70, z-score = -3.41, R) >droSec1.super_14 1695584 95 - 2068291 CAUAAAGAUUAUGAAAAGUCGUUAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCU-GAGCU----------------------GGUA-CG-AAAAUGGCUCAAAUAAAAUAAAACAG .....(((((((((...(((((....................)))))..)))))))))-(((((----------------------(...-..-....))))))................ ( -19.45, z-score = -2.99, R) >droYak2.chr2L 1732714 96 - 22324452 CAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCUGGAGCU----------------------GGUA-CG-AAAAUGGCUCAAAUAAAAUAAAACAG .....(((((((((...((((((..................))))))..))))))))).(((((----------------------(...-..-....))))))................ ( -20.07, z-score = -3.24, R) >droEre2.scaffold_4929 1805420 95 - 26641161 CAUAAAGAUUAUGAGAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCU-GAGCU----------------------GGUA-CG-AAAAUGGCUCAAAUAAAAUGAAACAG (((..(((((((((...((((((..................))))))..)))))))))-(((((----------------------(...-..-....)))))).......)))...... ( -19.17, z-score = -2.49, R) >droAna3.scaffold_12916 355676 96 + 16180835 CAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCGUAGUCU-GAGCU----------------------GGCGCCG-AAAAUGGCUCAAAUAAAAUAAAACAG .....(((((((((...((((((..................))))))..)))))))))-(((((----------------------(......-....))))))................ ( -19.57, z-score = -2.16, R) >dp4.chr4_group3 8487640 120 - 11692001 CAUAAAGAUUACAAAAAGUCGUAAAAGGCCAGAAAUAUGAAUGCGAUAGUCAUACUCUAGUACUAGUCCUCGUUCUCCGCUGCGAGGGCACCGAAAAAUGGCUCAAAUAAAAUAAAACAG ......((((......))))......(((((....(((((.........)))))...........((((((((........)))))))).........)))))................. ( -23.10, z-score = -1.25, R) >droPer1.super_1 9937622 120 - 10282868 CAUAAAGAUUACAAAAAGUCGUAAAAGGCCAGAAAUAUGAAUGCGAUAGUCAUACUCUAGUACUAGUCCUCGUUCUCCGCUGCGAGGGCACCGAAAAAUGGCUUAAAUAAAAUAAAACAG ......((((......)))).....((((((....(((((.........)))))...........((((((((........)))))))).........))))))................ ( -24.30, z-score = -1.63, R) >consensus CAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCU_GAGCU______________________GGUA_CG_AAAAUGGCUCAAAUAAAAUAAAACAG .....(((((((((...((((((..................))))))..))))))))).(((((...................................)))))................ (-12.17 = -13.36 + 1.19)
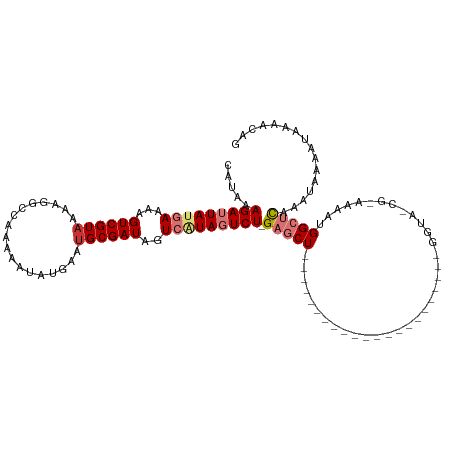
| Location | 1,761,127 – 1,761,228 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.22 |
| Shannon entropy | 0.46014 |
| G+C content | 0.48535 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -11.17 |
| Energy contribution | -12.22 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.666477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1761127 101 - 23011544 UCCGGCGCCACCCACCGCCCACCGCCUGCCGCCCAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCU-GAGCUGGUAC----------- ...((((........)))).......((((((......(((((((((...((((((..................))))))..)))))))))-..)).)))).----------- ( -23.97, z-score = -1.44, R) >droSim1.chr2L 1661296 93 - 22036055 CCCGGCGCCACCCACCGCCC--------CCGCCCAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUUUGAAUGCGAUAGUCAUAGUCU-GAGCUGGUAC----------- ...((((........)))).--------..(((.....(((((((((...((((((.(((........)))...))))))..)))))))))-.....)))..----------- ( -23.80, z-score = -2.57, R) >droSec1.super_14 1695611 93 - 2068291 CCCGGCGCCACCCACCGCCA--------CCGCCCAUAAAGAUUAUGAAAAGUCGUUAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCU-GAGCUGGUAC----------- ...((((........)))).--------..(((.....(((((((((...(((((....................)))))..)))))))))-.....)))..----------- ( -23.45, z-score = -2.21, R) >droYak2.chr2L 1732741 93 - 22324452 CCCGGCGCCAGCCGCCCAC---------CGCCCCAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCUGGAGCUGGUAC----------- ..((((....))))...((---------(((.(((....((((((((...((((((..................))))))..))))))))))).)).)))..----------- ( -26.97, z-score = -2.80, R) >droEre2.scaffold_4929 1805447 100 - 26641161 CCCGGCGCCGCCCAUCGCCCACCGCCC-ACGCCCAUAAAGAUUAUGAGAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCU-GAGCUGGUAC----------- ...((((..((.....))....)))).-..(((.....(((((((((...((((((..................))))))..)))))))))-.....)))..----------- ( -22.67, z-score = -0.81, R) >droAna3.scaffold_12916 355703 97 + 16180835 --CUCUACCGCCCUCUACCCAUUUA---CCGCCCAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCGUAGUCU-GAGCUGGCGCC---------- --.......................---.((((.....(((((((((...((((((..................))))))..)))))))))-.....))))..---------- ( -20.97, z-score = -1.72, R) >dp4.chr4_group3 8487680 99 - 11692001 ---------GGUCUGAGCCAAAA-----CCCCCCAUAAAGAUUACAAAAAGUCGUAAAAGGCCAGAAAUAUGAAUGCGAUAGUCAUACUCUAGUACUAGUCCUCGUUCUCCGC ---------((((((.(((....-----.....(.....).((((........))))..))))))).....(((((.(.(((((........).))))...).))))).)).. ( -15.00, z-score = -0.48, R) >droPer1.super_1 9937662 99 - 10282868 ---------GGUCUGAGCCAAAA-----CCCCCCAUAAAGAUUACAAAAAGUCGUAAAAGGCCAGAAAUAUGAAUGCGAUAGUCAUACUCUAGUACUAGUCCUCGUUCUCCGC ---------((((((.(((....-----.....(.....).((((........))))..))))))).....(((((.(.(((((........).))))...).))))).)).. ( -15.00, z-score = -0.48, R) >consensus CCCGGCGCCACCCACCGCCCA_______CCGCCCAUAAAGAUUAUGAAAAGUCGUAAAAGGCCAAAAAUAUGAAUGCGAUAGUCAUAGUCU_GAGCUGGUAC___________ ................................(((...(((((((((...((((((..................))))))..))))))))).....))).............. (-11.17 = -12.22 + 1.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:04 2011