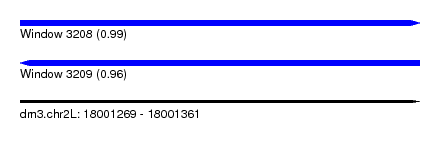
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 18,001,269 – 18,001,361 |
| Length | 92 |
| Max. P | 0.989672 |

| Location | 18,001,269 – 18,001,361 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 67.40 |
| Shannon entropy | 0.69089 |
| G+C content | 0.38797 |
| Mean single sequence MFE | -20.91 |
| Consensus MFE | -12.05 |
| Energy contribution | -12.08 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.989672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
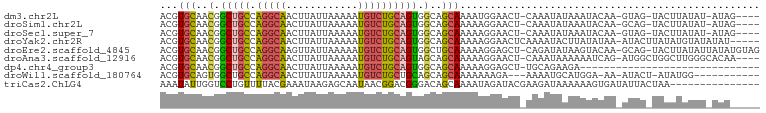
>dm3.chr2L 18001269 92 + 23011544 ACGUGCAACGGCUGCCAGGCAACUUAUUAAAAAUGUCUGCAGUGGCAGCAAAAUGGAACU-CAAAUAUAAAUACAA-GUAG-UACUUAUAU-AUAG---- .((.....))(((((((.(((((...........)).)))..)))))))...........-...((((((.(((..-...)-)).))))))-....---- ( -21.10, z-score = -1.48, R) >droSim1.chr2L 17682289 92 + 22036055 ACGUGCAACGGCUGCCAGGCAACUUAUUAAAAAUGUCUGCAGUGGCAGCAAAAAGGAACU-CAAAUAUAAAUACAA-GCAG-UACUUAUAU-AUAG---- .((.....))(((((((.(((((...........)).)))..)))))))...........-...((((((.(((..-...)-)).))))))-....---- ( -21.10, z-score = -1.59, R) >droSec1.super_7 1636243 92 + 3727775 ACGUGCAACGGCUGCCAGGCAACUUAUUAAAAAUGUCUGCAGUGGCAGCAAAAAGGAACU-CAAAUAUAAAUACAA-GUAG-UACUUAUAU-AUAG---- .((.....))(((((((.(((((...........)).)))..)))))))...........-...((((((.(((..-...)-)).))))))-....---- ( -21.10, z-score = -1.72, R) >droYak2.chr2R 18047462 94 - 21139217 ACGUGCAACGGCUGCCAGGCAACUUAUUAAAAAUGUCUGCAGUGGCAGCAAAAAGGAACUCAAAAUACUUAUAUAA-AUACUUAUAUGUAUAUAU----- .((.....))(((((((.(((((...........)).)))..)))))))...............((((.((((...-.....)))).))))....----- ( -20.40, z-score = -1.51, R) >droEre2.scaffold_4845 16325207 97 - 22589142 ACGUGCAACGGCUGCCAGGCAAGUUAUUAAAAAUGUCUGCAGUGGCUGCAAAAAGGAGCU-CAGAUAUAAGUACAA-GCAG-UACUUAUAUUAUAUGUAG (((((((.(.(((((.(((((..((....))..)))))))))).).)))...........-..(((((((((((..-...)-))))))))))..)))).. ( -30.20, z-score = -2.64, R) >droAna3.scaffold_12916 13281985 94 - 16180835 ACGUGCAACGGCUGCCAGGCAACUUAUUAAAAAUGUCUGCAGUAGCAGCAAAAAGGAACU-CAAAUAAAAAAUCAG-AUGGCUGGCUUGGGCACAA---- ..((((...(((((((((....)..........((.((((....))))))..........-...............-.)))).))))...))))..---- ( -24.60, z-score = -0.98, R) >dp4.chr4_group3 1312079 69 - 11692001 ACGUGCAACGGCUGCCAGGCAACUUAUUAAAAAUGUCUGCAGUGGCAGCAAAAAGGAGCU-UGCAGAAGA------------------------------ ...((((((.(((((((.(((((...........)).)))..)))))))........).)-)))).....------------------------------ ( -19.20, z-score = -0.17, R) >droWil1.scaffold_180764 3550893 83 - 3949147 ACGUGCAGUGGCUGCCAGGCAACUUAUUAAAAAUGUCUGCUGCAGCAGCAAAAAAAGA---AAAAUGCAUGGA-AA-AUACU-AUAUGG----------- .((((((((.(((((((((((............))))))..))))).)).........---....))))))..-..-.....-......----------- ( -20.01, z-score = -0.91, R) >triCas2.ChLG4 4741396 85 + 13894384 AAAUAUUGGUCCUGUUUUACGAAAUAAGAGCAAUAACGGACGGGACAGCAAAAUAGAUACGAAGAUAAAAAAGUGAUAUUACUAA--------------- ........(((((((((....................))))))))).........................((((....))))..--------------- ( -10.45, z-score = -0.49, R) >consensus ACGUGCAACGGCUGCCAGGCAACUUAUUAAAAAUGUCUGCAGUGGCAGCAAAAAGGAACU_CAAAUAUAAAUACAA_AUAG_UACUUAUAU_AUA_____ ...(((..(.(((((.(((((............)))))))))).)..))).................................................. (-12.05 = -12.08 + 0.03)
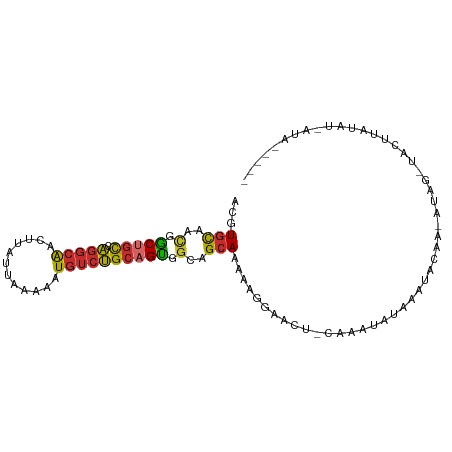
| Location | 18,001,269 – 18,001,361 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 67.40 |
| Shannon entropy | 0.69089 |
| G+C content | 0.38797 |
| Mean single sequence MFE | -18.87 |
| Consensus MFE | -10.42 |
| Energy contribution | -10.77 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 18001269 92 - 23011544 ----CUAU-AUAUAAGUA-CUAC-UUGUAUUUAUAUUUG-AGUUCCAUUUUGCUGCCACUGCAGACAUUUUUAAUAAGUUGCCUGGCAGCCGUUGCACGU ----((..-(((((((((-(...-..))))))))))...-)).........(((((((..((((...((......)).)))).))))))).......... ( -23.10, z-score = -2.20, R) >droSim1.chr2L 17682289 92 - 22036055 ----CUAU-AUAUAAGUA-CUGC-UUGUAUUUAUAUUUG-AGUUCCUUUUUGCUGCCACUGCAGACAUUUUUAAUAAGUUGCCUGGCAGCCGUUGCACGU ----((..-(((((((((-(...-..))))))))))...-)).........(((((((..((((...((......)).)))).))))))).......... ( -23.10, z-score = -1.88, R) >droSec1.super_7 1636243 92 - 3727775 ----CUAU-AUAUAAGUA-CUAC-UUGUAUUUAUAUUUG-AGUUCCUUUUUGCUGCCACUGCAGACAUUUUUAAUAAGUUGCCUGGCAGCCGUUGCACGU ----((..-(((((((((-(...-..))))))))))...-)).........(((((((..((((...((......)).)))).))))))).......... ( -23.10, z-score = -2.34, R) >droYak2.chr2R 18047462 94 + 21139217 -----AUAUAUACAUAUAAGUAU-UUAUAUAAGUAUUUUGAGUUCCUUUUUGCUGCCACUGCAGACAUUUUUAAUAAGUUGCCUGGCAGCCGUUGCACGU -----.((((((.(((....)))-.))))))....................(((((((..((((...((......)).)))).))))))).......... ( -17.10, z-score = -0.13, R) >droEre2.scaffold_4845 16325207 97 + 22589142 CUACAUAUAAUAUAAGUA-CUGC-UUGUACUUAUAUCUG-AGCUCCUUUUUGCAGCCACUGCAGACAUUUUUAAUAACUUGCCUGGCAGCCGUUGCACGU .........(((((((((-(...-..))))))))))...-..........((((((..((((((.((..((....))..)).)).))))..))))))... ( -23.80, z-score = -1.86, R) >droAna3.scaffold_12916 13281985 94 + 16180835 ----UUGUGCCCAAGCCAGCCAU-CUGAUUUUUUAUUUG-AGUUCCUUUUUGCUGCUACUGCAGACAUUUUUAAUAAGUUGCCUGGCAGCCGUUGCACGU ----..((((....(((((.((.-(((((((.......)-))))......((((((....)))).)).........)).)).))))).......)))).. ( -20.20, z-score = -0.16, R) >dp4.chr4_group3 1312079 69 + 11692001 ------------------------------UCUUCUGCA-AGCUCCUUUUUGCUGCCACUGCAGACAUUUUUAAUAAGUUGCCUGGCAGCCGUUGCACGU ------------------------------.....((((-(..........(((((((..((((...((......)).)))).)))))))..)))))... ( -16.10, z-score = -0.11, R) >droWil1.scaffold_180764 3550893 83 + 3949147 -----------CCAUAU-AGUAU-UU-UCCAUGCAUUUU---UCUUUUUUUGCUGCUGCAGCAGACAUUUUUAAUAAGUUGCCUGGCAGCCACUGCACGU -----------......-.((((-..-...)))).....---.........(((((((((((...............)))))..)))))).......... ( -14.76, z-score = 0.37, R) >triCas2.ChLG4 4741396 85 - 13894384 ---------------UUAGUAAUAUCACUUUUUUAUCUUCGUAUCUAUUUUGCUGUCCCGUCCGUUAUUGCUCUUAUUUCGUAAAACAGGACCAAUAUUU ---------------.((((((...........................))))))....(((((((.((((.........))))))).))))........ ( -8.53, z-score = -0.71, R) >consensus _____UAU_AUAUAAGUA_CUAC_UUGUAUUUAUAUUUG_AGUUCCUUUUUGCUGCCACUGCAGACAUUUUUAAUAAGUUGCCUGGCAGCCGUUGCACGU ...................................................(((((((..((((...((......)).)))).))))))).......... (-10.42 = -10.77 + 0.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:12 2011