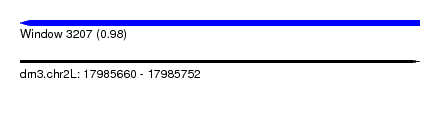
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,985,660 – 17,985,752 |
| Length | 92 |
| Max. P | 0.982210 |

| Location | 17,985,660 – 17,985,752 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 71.21 |
| Shannon entropy | 0.47032 |
| G+C content | 0.47174 |
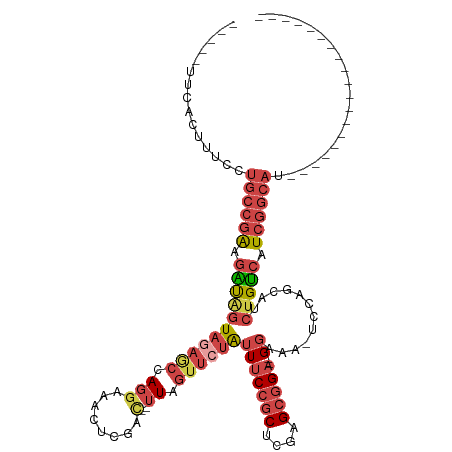
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -18.09 |
| Energy contribution | -19.40 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.982210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
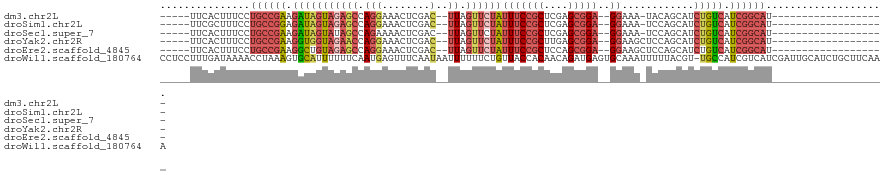
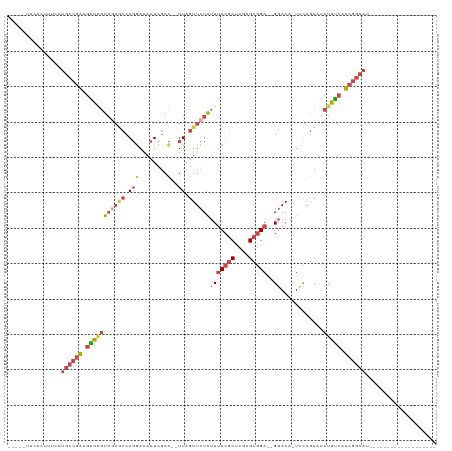
>dm3.chr2L 17985660 92 - 23011544 -----UUCACUUUCCUGCCGAAGAUAGUAGAGCCAGGAAACUCGAC--UUAGUUCUAUUUCCGCUCGAGCGGA--GGAAA-UACAGCAUCUGUCAUCGGCAU------------------- -----..........((((((.(((((((((((.(((........)--)).))))))(((((((....)))))--))...-........))))).)))))).------------------- ( -31.30, z-score = -3.03, R) >droSim1.chr2L 17666543 92 - 22036055 -----UUCGCUUUCCUGCCGGAGAUAGUAGAGCCAGGAAACUCGAC--UUAGUUCUAUUUCCGCUCGAGCGGA--GGAAA-UCCAGCAUCUGUCAUCGGCAU------------------- -----..........(((((..(((((((((((.(((........)--)).))))))..(((((....)))))--((...-.)).....)))))..))))).------------------- ( -30.60, z-score = -1.56, R) >droSec1.super_7 1620526 92 - 3727775 -----UUCACUUUCCUGCCGAAGAUAGUAUAGCCAGAAAACUCGAC--UUAGUUCUAUUUCCGCUCGAGCGGA--GGAAA-UCCAGCAUCUGUCAUCGGCAU------------------- -----..........((((((.(((((.((.((..((.((((....--..))))...(((((((....)))))--))...-))..))))))))).)))))).------------------- ( -27.30, z-score = -2.43, R) >droYak2.chr2R 18028103 93 + 21139217 -----UUCACUUUCCUGCCGAAGGUGGUAGAACCAGGAAACUCGAC--UUAGUUCUAUUUCCGCUUGAGCGGA--GGAAGCUCCAGCAUCUGUCAUCGGCAU------------------- -----..........((((((.((..(((((((.(((........)--)).)))))))..))(((.((((...--....)))).)))........)))))).------------------- ( -32.90, z-score = -2.39, R) >droEre2.scaffold_4845 16309104 93 + 22589142 -----UUCACUUUCCUGCCGAAGGCUGUAGAGCCAGGAAACUCGAC--UUAGUUCUAUUUCCGCUCCAGCGGA--GGAAGCUCCAGCAUCUGUCAUCGGCAU------------------- -----..........((((((.((((....))))((....)).(((--...........(((((....)))))--(((.((....)).)))))).)))))).------------------- ( -31.70, z-score = -1.95, R) >droWil1.scaffold_180764 412377 120 - 3949147 CCUCCUUUGAUAAAACCUAAAGUGCAUUUUUUCAAUGAGUUUCAAUAAUUUUUUCUGUUACCACAACAGAUGAGUGCAAAUUUUUACGU-UGCCAUCGUCAUCGAUUGCAUCUGCUUCAAA ......((((...(((.((((((((((((......((.....)).........((((((.....)))))).)))))))...))))).))-)((.((((....)))).)).......)))). ( -19.40, z-score = -0.74, R) >consensus _____UUCACUUUCCUGCCGAAGAUAGUAGAGCCAGGAAACUCGAC__UUAGUUCUAUUUCCGCUCGAGCGGA__GGAAA_UCCAGCAUCUGUCAUCGGCAU___________________ ...............((((((.(((((.......((....))..........((((...(((((....)))))..))))..........))))).)))))).................... (-18.09 = -19.40 + 1.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:10 2011