
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,972,379 – 17,972,499 |
| Length | 120 |
| Max. P | 0.735258 |

| Location | 17,972,379 – 17,972,499 |
|---|---|
| Length | 120 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.47 |
| Shannon entropy | 0.36608 |
| G+C content | 0.57121 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -23.03 |
| Energy contribution | -23.21 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735258 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
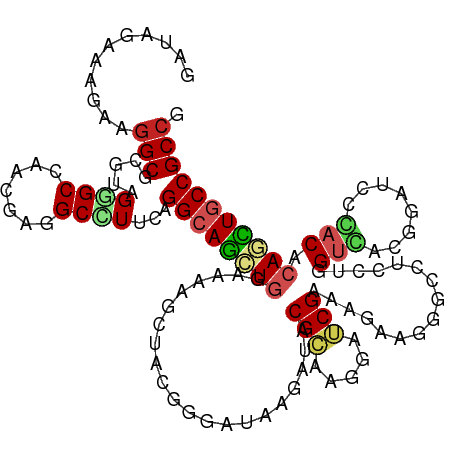
>dm3.chr2L 17972379 120 + 23011544 GAUAGGAAAAAGGCUCGUAGGGCCAACGAGGCCUUCAGGCAGCUGGAAAAGCUAAGGGAUAAGAUCCGAAAGGAUCGAAAGAAGGGCUUGCUUGUCACGGAUCCGACGCAGCUGCCGCCG ...........(((..(.((((((.....))))))).((((((((...(((((.........(((((....)))))........)))))....(((........))).))))))))))). ( -49.93, z-score = -3.40, R) >droSim1.chr2L 17650841 120 + 22036055 GAUAGGAAGAAGGCUCGUAGGGCCAACGAGGCCUUCAGGCAGCUGGAAAAGCUAAGGGAUAAGAUCCGAAAAGAUCGAAAGAAGGGCUUGCUUGUCACGGAUCCGACGCAGCUGCCGCCG ....((..((((((((((.......)))).)))))).((((((((((.((((.(((((((...)))).......((....))....))))))).)).((....))...)))))))).)). ( -46.30, z-score = -2.44, R) >droSec1.super_7 1607276 120 + 3727775 GAUAGGAAGAAGGCUCGUAGGGCCAACGAGGCCUUCAGGCAGCUGGAAAAGCUAAGGGAUAAGAUCCGAAAGGAUCGAAAGAAGGGCUUGCUUGUCACGGAUCCGACGCAGCUGCCGCCG ....((..((((((((((.......)))).)))))).((((((((...(((((.........(((((....)))))........)))))....(((........))).)))))))).)). ( -51.03, z-score = -3.52, R) >droYak2.chr2R 18015212 120 - 21139217 GAUAGAAAGAAGGCUCGUCGAGCCAACGAGGCCUUCAGGCAGCUGGAAAAGCUAAGGGAUAAGAUCCGAAAGGAUCGAAAGAAGGGCUUGCUUGUCACGGAUCCGACCCAGCUGCCGCCG ........((((((((((.......)))).)))))).(((((((((..(((((.........(((((....)))))........)))))........((....))..))))))))).... ( -51.13, z-score = -3.78, R) >droEre2.scaffold_4845 16294959 120 - 22589142 GAUAGAAAGAAGGCGCGUAGAGCCAACGAGGCCUUCAGGCAGCUGGAAAAGCUAAGGGAUAAGAUCCGAAAGGAUCGAAAGAAGGGCUUGCUUGUCACGGAUCCAACGCAGCUGCCGCCG ........((((((.(((.......)))..)))))).((((((((((.(((((.........(((((....)))))........))))).....)).((.......)))))))))).... ( -45.63, z-score = -2.80, R) >dp4.chr4_group3 1281054 120 - 11692001 GACAGAAAGAAGGCGCGGAGGGCGAACGAGGCCUUCCGGCAGCUCGAGAAGCUGCGCGACAAGAUACGCAAGGACCGCAAGAAGGGACUCCUGGUGACCGAUCCCUCACAGCUCCCGCCC ...........((((((((((((.......))))))))((((((.....))))))(((........)))..(((..((....(((((.((..(....).)))))))....))))))))). ( -45.00, z-score = -0.92, R) >droPer1.super_8 2342200 120 - 3966273 GACAGAAAGAAGGCGCGGAGGGCGAACGAGGCCUUCCGGCAGCUCGAGAAGCUGCGCGACAAGAUACGCAAGGACCGCAAGAAGGGACUCCUGGUGACCGAUCCCUCACAGCUCCCGCCC ...........((((((((((((.......))))))))((((((.....))))))(((........)))..(((..((....(((((.((..(....).)))))))....))))))))). ( -45.00, z-score = -0.92, R) >droWil1.scaffold_180764 3516351 120 - 3949147 GAUCGUAAAAAGGCCAGAAGAGCGAAUGAGGCCUUCCGGCAAUUGGAAAAGUUACGUGACAAAAUACGCAAAGAGCGGAAAAAGGGUCUCUUGGUAACAGAUCCCACACAAUUGCCGCCG ...........(((..((((.((.......)))))).(((((((((........)(((........(((.....)))......((((((.........))))))))).))))))))))). ( -32.30, z-score = -1.31, R) >droMoj3.scaffold_6500 22055376 120 + 32352404 GAUAGAAAGAAGGCGCGACGCGCCAAUGAGGCGUUCCGGCAGCUGGAGAAGCUACGUGAUAAGAUACGCAAGGAGCGCAAGAAGGGUCUCCUGGUGACUGAUCCCACACAACUGCCGCCC ...........((((.((.(((((.....))))))))(((((((.....))))..(((....(((.(((.((((((.(.....).).))))).)))....)))...)))....)))))). ( -37.10, z-score = -0.16, R) >droGri2.scaffold_15252 14472686 120 + 17193109 GAUCGGAAGAAGGCGCGACGUGCGAAUGAGGCAUUCAGGCAGCUGGAGAAGCUGCGCGAUAAGAUACGCAAGGAGCGCAAGAAGGGACUCCUGGUGACGGAUGCUACGCAAUUGCCGCAU ..((....))..((((((..((((.....(((((((..((((((.....))))))(((........))).((((((.(.....).).)))))(....)))))))).)))).))).))).. ( -43.60, z-score = -2.03, R) >droVir3.scaffold_12963 17038579 120 + 20206255 GAUCGAAAGAAGGCGAGACGCGCCAACGAGGCGUUCCGGCAGCUGGAAAAGCUACGUGAUAAGAUACGCAAGGAGCGUAAGAAGGGUCUCCUGGUGACUGAUCCCACACAAUUGCCGCCC ..((....)).((((.((.(((((.....))))))))(((((((.....))))..(((......(((((.....)))))....(((((....(....).)))))..)))....)))))). ( -42.20, z-score = -2.06, R) >consensus GAUAGAAAGAAGGCGCGUAGGGCCAACGAGGCCUUCAGGCAGCUGGAAAAGCUACGGGAUAAGAUACGCAAGGAUCGAAAGAAGGGCCUCCUGGUCACGGAUCCCACACAGCUGCCGCCG ...........(((.....((((.......))))...((((((((.....................(((.....)))................(((........))).))))))))))). (-23.03 = -23.21 + 0.17)


| Location | 17,972,379 – 17,972,499 |
|---|---|
| Length | 120 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.47 |
| Shannon entropy | 0.36608 |
| G+C content | 0.57121 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -26.16 |
| Energy contribution | -25.09 |
| Covariance contribution | -1.07 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17972379 120 - 23011544 CGGCGGCAGCUGCGUCGGAUCCGUGACAAGCAAGCCCUUCUUUCGAUCCUUUCGGAUCUUAUCCCUUAGCUUUUCCAGCUGCCUGAAGGCCUCGUUGGCCCUACGAGCCUUUUUCCUAUC .(((((((((((.((((......))))(((((((.....)))..(((((....)))))..........))))...))))))))....((((.....))))......)))........... ( -39.80, z-score = -1.95, R) >droSim1.chr2L 17650841 120 - 22036055 CGGCGGCAGCUGCGUCGGAUCCGUGACAAGCAAGCCCUUCUUUCGAUCUUUUCGGAUCUUAUCCCUUAGCUUUUCCAGCUGCCUGAAGGCCUCGUUGGCCCUACGAGCCUUCUUCCUAUC .((.((((((((.((((......))))(((((((.......(((((.....)))))........))).))))...)))))))).((((((.((((.......))))))))))..)).... ( -40.46, z-score = -2.08, R) >droSec1.super_7 1607276 120 - 3727775 CGGCGGCAGCUGCGUCGGAUCCGUGACAAGCAAGCCCUUCUUUCGAUCCUUUCGGAUCUUAUCCCUUAGCUUUUCCAGCUGCCUGAAGGCCUCGUUGGCCCUACGAGCCUUCUUCCUAUC .((.((((((((.((((......))))(((((((.....)))..(((((....)))))..........))))...)))))))).((((((.((((.......))))))))))..)).... ( -41.10, z-score = -2.27, R) >droYak2.chr2R 18015212 120 + 21139217 CGGCGGCAGCUGGGUCGGAUCCGUGACAAGCAAGCCCUUCUUUCGAUCCUUUCGGAUCUUAUCCCUUAGCUUUUCCAGCUGCCUGAAGGCCUCGUUGGCUCGACGAGCCUUCUUUCUAUC ....(((((((((((((......))))(((((((.....)))..(((((....)))))..........))))..))))))))).((((((.((((((...))))))))))))........ ( -46.20, z-score = -2.76, R) >droEre2.scaffold_4845 16294959 120 + 22589142 CGGCGGCAGCUGCGUUGGAUCCGUGACAAGCAAGCCCUUCUUUCGAUCCUUUCGGAUCUUAUCCCUUAGCUUUUCCAGCUGCCUGAAGGCCUCGUUGGCUCUACGCGCCUUCUUUCUAUC .(((((((((((....(....)..((.(((((((.....)))..(((((....)))))..........)))).)))))))))).(.(((((.....))).)).)..)))........... ( -35.50, z-score = -0.70, R) >dp4.chr4_group3 1281054 120 + 11692001 GGGCGGGAGCUGUGAGGGAUCGGUCACCAGGAGUCCCUUCUUGCGGUCCUUGCGUAUCUUGUCGCGCAGCUUCUCGAGCUGCCGGAAGGCCUCGUUCGCCCUCCGCGCCUUCUUUCUGUC (((((((((((((.((((((((.....((((((....))))))))))))))(((........))))))))))).((((..(((....)))))))..)))))................... ( -49.80, z-score = -0.73, R) >droPer1.super_8 2342200 120 + 3966273 GGGCGGGAGCUGUGAGGGAUCGGUCACCAGGAGUCCCUUCUUGCGGUCCUUGCGUAUCUUGUCGCGCAGCUUCUCGAGCUGCCGGAAGGCCUCGUUCGCCCUCCGCGCCUUCUUUCUGUC (((((((((((((.((((((((.....((((((....))))))))))))))(((........))))))))))).((((..(((....)))))))..)))))................... ( -49.80, z-score = -0.73, R) >droWil1.scaffold_180764 3516351 120 + 3949147 CGGCGGCAAUUGUGUGGGAUCUGUUACCAAGAGACCCUUUUUCCGCUCUUUGCGUAUUUUGUCACGUAACUUUUCCAAUUGCCGGAAGGCCUCAUUCGCUCUUCUGGCCUUUUUACGAUC ((.(((((((((...(((.(((.......)))..)))............((((((........))))))......)))))))))(((((((..............)))))))...))... ( -34.04, z-score = -1.83, R) >droMoj3.scaffold_6500 22055376 120 - 32352404 GGGCGGCAGUUGUGUGGGAUCAGUCACCAGGAGACCCUUCUUGCGCUCCUUGCGUAUCUUAUCACGUAGCUUCUCCAGCUGCCGGAACGCCUCAUUGGCGCGUCGCGCCUUCUUUCUAUC (((((((((((...(((((....)).)))(((((..((...(((((.....)))))...........))..))))))))))))((..((((.....))))..))..)))).......... ( -41.34, z-score = -1.05, R) >droGri2.scaffold_15252 14472686 120 - 17193109 AUGCGGCAAUUGCGUAGCAUCCGUCACCAGGAGUCCCUUCUUGCGCUCCUUGCGUAUCUUAUCGCGCAGCUUCUCCAGCUGCCUGAAUGCCUCAUUCGCACGUCGCGCCUUCUUCCGAUC ..(((((...((((((((....))....((((((.(......).)))))))))))).......((((((((.....))))))..(((((...)))))))..))))).............. ( -33.40, z-score = -0.92, R) >droVir3.scaffold_12963 17038579 120 - 20206255 GGGCGGCAAUUGUGUGGGAUCAGUCACCAGGAGACCCUUCUUACGCUCCUUGCGUAUCUUAUCACGUAGCUUUUCCAGCUGCCGGAACGCCUCGUUGGCGCGUCUCGCCUUCUUUCGAUC (((((((((..((((((((...(((.......)))...))))))))...))))............((((((.....)))))).((..((((.....))))..)).))))).......... ( -37.90, z-score = -0.68, R) >consensus CGGCGGCAGCUGCGUGGGAUCCGUCACCAGGAGGCCCUUCUUUCGAUCCUUGCGUAUCUUAUCCCGUAGCUUUUCCAGCUGCCUGAAGGCCUCGUUGGCCCUACGCGCCUUCUUUCUAUC .(((((((((((.((((......))))..............(((((.....)))))...................))))))))....(((.......)))......)))........... (-26.16 = -25.09 + -1.07)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:09 2011