| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,911,127 – 17,911,237 |
| Length | 110 |
| Max. P | 0.984982 |

| Location | 17,911,127 – 17,911,237 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 59.70 |
| Shannon entropy | 0.56429 |
| G+C content | 0.30269 |
| Mean single sequence MFE | -15.03 |
| Consensus MFE | -7.56 |
| Energy contribution | -8.90 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.984982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17911127 110 - 23011544 AUUAAAUCAAAUACUCAUUACACAAUUUA-UUACAUUUUCAGAGAAUGCACGUGUUUUCCCAUUUUAAUAUAAUCCACAUUAAAAAGAACAUUCACAGGCA--AAUAAGCCCA .............................-...(((((.....)))))...(((((((....(((((((.........)))))))))))))).....(((.--.....))).. ( -8.80, z-score = -0.68, R) >droEre2.scaffold_4845 16237932 88 + 22589142 ------------------------AUUAU-UUACAUUUUCAGAGAAGGCAAGGGUUUUCUCAUUUCAGUGUAAUCCACAUUGAAAAGAACAUUCUCAGGCAACAAAAAGCUUA ------------------------.....-.......(((.((((((((....)))))))).(((((((((.....))))))))).))).......((((........)))). ( -20.30, z-score = -3.55, R) >droYak2.chr2R 17951967 91 + 21139217 ------AUUAAUGUUCACUUUGCAAUUUUAUUAAAUGUCCAGAGAAGGCAAGUGUUUCCUCAUUUCCAUAGAAUCCAUAUUUAAAUGAACAUUCUCA---------------- ------...(((((((((((((..((((....))))...)))))..((.(((((......)))))))..................))))))))....---------------- ( -16.00, z-score = -1.94, R) >consensus _________AAU__UCA_U___CAAUUUU_UUACAUUUUCAGAGAAGGCAAGUGUUUUCUCAUUUCAAUAUAAUCCACAUUAAAAAGAACAUUCUCAGGCA__AA_AAGC__A .........................................((((((((....)))))))).((((((((.......))))))))............................ ( -7.56 = -8.90 + 1.34)


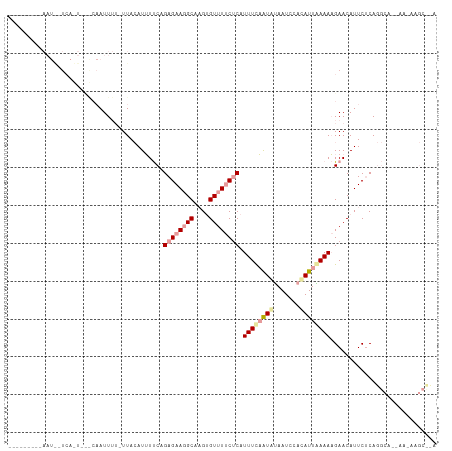
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:02 2011