| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,760,993 – 1,761,067 |
| Length | 74 |
| Max. P | 0.998603 |

| Location | 1,760,993 – 1,761,067 |
|---|---|
| Length | 74 |
| Sequences | 7 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 62.15 |
| Shannon entropy | 0.74647 |
| G+C content | 0.51656 |
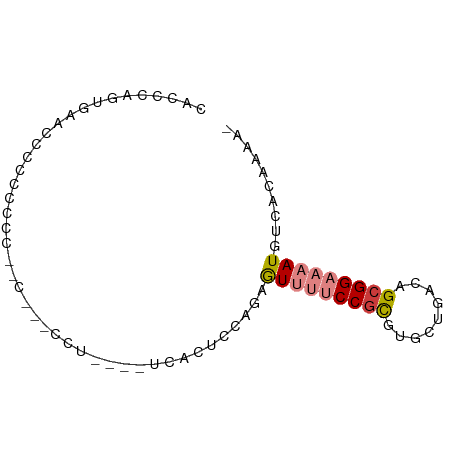
| Mean single sequence MFE | -16.47 |
| Consensus MFE | -8.42 |
| Energy contribution | -9.06 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
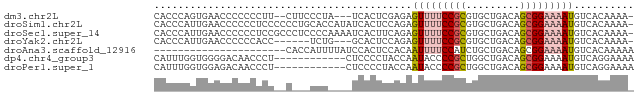
>dm3.chr2L 1760993 74 + 23011544 CACCCAGUGAACCCCCCCUU--CUUCCCUA---UCACUCGAGAGUUUUCCGCGUGCUGACAGCGGAAAAUGUCACAAAA- ......((((.........(--(((.....---......))))(((((((((((....)).))))))))).))))....- ( -16.30, z-score = -2.05, R) >droSim1.chr2L 1661157 79 + 22036055 CACCCAUUGAACCCCCCUCCCCCCUGCACCAUAUCACUCCAGAGUUUUCCGCGUGCUGACAGCGGAAAAUGUCACAAAA- .........................................(((((((((((((....)).))))))))).))......- ( -12.60, z-score = -1.23, R) >droSec1.super_14 1695472 79 + 2068291 CACCCAUUGAACCCCCCUCCGCCCUCCCCAAAAUCACUUCAGAGUUUUCCGCGUGCUGACAGCGGAAAAUGUCACAAAA- .........................................(((((((((((((....)).))))))))).))......- ( -12.60, z-score = -1.22, R) >droYak2.chr2L 1732611 70 + 22324452 CACCCAUUGAACCCCCCACC------UCUG---GCACUCCAGAGUUUUCCGCGUGCUGACAGCGGAAAAUGUCACAAAA- .......(((.........(------((((---(....))))))((((((((((....)).))))))))..))).....- ( -18.80, z-score = -2.46, R) >droAna3.scaffold_12916 355585 58 - 16180835 ----------------------CACCAUUUUAUCCACUCCACAAUUUUCCAUCUGCUGACAGCGGAAAAUGUCACAAAAA ----------------------.....................(((((((..(((....))).))))))).......... ( -7.50, z-score = -1.37, R) >dp4.chr4_group3 8487539 68 + 11692001 CAUUUGGUGGGGACAACCCU------------CUCCCCUACCAAUACCCCGCUGGCUGACAGCGGAAAAUGUCAGGAAAA ...((((((((((.......------------.))))).)))))....((((((.....))))))............... ( -25.50, z-score = -2.68, R) >droPer1.super_1 9937521 68 + 10282868 CAUUUGGUGGAGACAACCCU------------CUCCCCUACCAAUACCCCGCUGGCUGACAGCGGAAAAUGUCAGGAAAA (((((((.(((((......)------------))))))..........((((((.....)))))).)))))......... ( -22.00, z-score = -2.49, R) >consensus CACCCAGUGAACCCCCCCCC__C___CCU____UCACUCCAGAGUUUUCCGCGUGCUGACAGCGGAAAAUGUCACAAAA_ ...........................................(((((((((.........))))))))).......... ( -8.42 = -9.06 + 0.63)
| Location | 1,760,993 – 1,761,067 |
|---|---|
| Length | 74 |
| Sequences | 7 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 62.15 |
| Shannon entropy | 0.74647 |
| G+C content | 0.51656 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -8.34 |
| Energy contribution | -8.53 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.588863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1760993 74 - 23011544 -UUUUGUGACAUUUUCCGCUGUCAGCACGCGGAAAACUCUCGAGUGA---UAGGGAAG--AAGGGGGGGUUCACUGGGUG -....((((..((((((((((....)).))))))))(((((......---........--..)))))...))))...... ( -19.19, z-score = 0.11, R) >droSim1.chr2L 1661157 79 - 22036055 -UUUUGUGACAUUUUCCGCUGUCAGCACGCGGAAAACUCUGGAGUGAUAUGGUGCAGGGGGGAGGGGGGUUCAAUGGGUG -..(((.(((.((..((.((((((.(((.((((....))))..)))...))..)))).))..))....))))))...... ( -20.40, z-score = -0.02, R) >droSec1.super_14 1695472 79 - 2068291 -UUUUGUGACAUUUUCCGCUGUCAGCACGCGGAAAACUCUGAAGUGAUUUUGGGGAGGGCGGAGGGGGGUUCAAUGGGUG -..(((.(((.(((((((((.((..(((.((((....))))..)))........)).)))))))))..))))))...... ( -21.40, z-score = -0.35, R) >droYak2.chr2L 1732611 70 - 22324452 -UUUUGUGACAUUUUCCGCUGUCAGCACGCGGAAAACUCUGGAGUGC---CAGA------GGUGGGGGGUUCAAUGGGUG -.....(((.(((..(((((....((((.((((....))))..))))---....------)))))..))))))....... ( -22.60, z-score = -1.00, R) >droAna3.scaffold_12916 355585 58 + 16180835 UUUUUGUGACAUUUUCCGCUGUCAGCAGAUGGAAAAUUGUGGAGUGGAUAAAAUGGUG---------------------- ..((..(..((((((((((((....))).))))))...)))..)..))..........---------------------- ( -12.70, z-score = -1.08, R) >dp4.chr4_group3 8487539 68 - 11692001 UUUUCCUGACAUUUUCCGCUGUCAGCCAGCGGGGUAUUGGUAGGGGAG------------AGGGUUGUCCCCACCAAAUG .............((((((((.....))))))))..(((((.(((((.------------.......))))))))))... ( -25.30, z-score = -1.65, R) >droPer1.super_1 9937521 68 - 10282868 UUUUCCUGACAUUUUCCGCUGUCAGCCAGCGGGGUAUUGGUAGGGGAG------------AGGGUUGUCUCCACCAAAUG ....((((.((..((((((((.....))))))))...)).))))((((------------(......)))))........ ( -23.90, z-score = -1.56, R) >consensus _UUUUGUGACAUUUUCCGCUGUCAGCACGCGGAAAACUCUGGAGUGA____AGG___G__AGGGGGGGGUUCAAUGGGUG ...........((((((((((....)).))))))))............................................ ( -8.34 = -8.53 + 0.19)

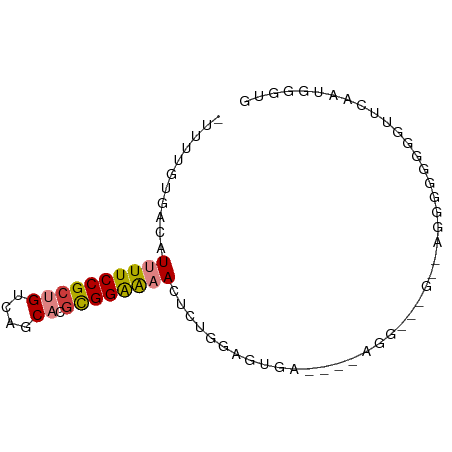
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:03 2011