
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,894,377 – 17,894,477 |
| Length | 100 |
| Max. P | 0.554140 |

| Location | 17,894,377 – 17,894,477 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 74.17 |
| Shannon entropy | 0.50992 |
| G+C content | 0.39469 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.23 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
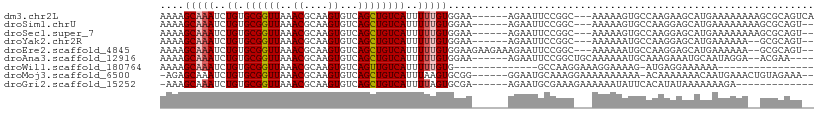
>dm3.chr2L 17894377 100 + 23011544 AAAAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGCUGUCAUUUUUGUGGAA------AGAAUUCCGGC---AAAAAGUGCCAAGAAGCAUGAAAAAAAAGCGCAGUCA ..........((((((......((....)).((((((.((........((((------....))))(((---(.....))))..))))).))).......))))))... ( -25.90, z-score = -1.54, R) >droSim1.chrU 13481056 98 - 15797150 AAAAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGCUGUCAUUUUUGUGGAA------AGAAUUCCGGC---AAAAAGUGCCAAGGAGCAUGAAAAAAAAGCGCAGU-- ..........((((((((((..((....))...)))).((((.(((.(((((------....))))(((---(.....)))).).))).)))).......)))))).-- ( -25.50, z-score = -1.38, R) >droSec1.super_7 1532940 98 + 3727775 AAAAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGCUGUCAUUUUUGUGGAA------AGAAUUCCGGC---AAAAAGUGCCAAGGAGCAUGAAAAAAAAGCGCAGU-- ..........((((((((((..((....))...)))).((((.(((.(((((------....))))(((---(.....)))).).))).)))).......)))))).-- ( -25.50, z-score = -1.38, R) >droYak2.chr2R 17934841 96 - 21139217 AAAAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGCUGUCAUUUUUGUGGAA------AGAAUUCCGGC---AAAAAAUGCCAAGGAGCAUGAAAAAA--GCGCAGU-- ..........((((((((((..((....))...)))).((((.(((.(((((------....))))(((---(.....)))).).))).)))).....--)))))).-- ( -25.50, z-score = -1.52, R) >droEre2.scaffold_4845 16221071 102 - 22589142 AAAAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGCUGUCAUUUUUGUGGAAGAAGAAAGAAUUCCGGC---AAAAAAUGCCAAGGAGCAUGAAAAAA--GCGCAGU-- ..........((((((((((..((....))...)))).((((.(((.(((((..........))))(((---(.....)))).).))).)))).....--)))))).-- ( -25.40, z-score = -1.31, R) >droAna3.scaffold_12916 13183798 97 - 16180835 AAAAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGCUGUCAUUUUUGUGGAA------AGAAUUCCGGCUGCAAAAAAUGCAAAGAAAUGCAAUAGGA--ACGAA---- ....(((..(((.((((.....((....))(.((((((..((((((.....)------)))))..)))))))......)))).)))..))).......--.....---- ( -24.70, z-score = -1.55, R) >droWil1.scaffold_180764 3419602 78 - 3949147 AAAAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGUUGUCAUUUUUGUG--------------GCCAAGGAAAGGAAAAG-AUGAGGAAAAAA---------------- ....(((......((((......))))..)))...((.((((((((...--------------.((.......)).))))-)))).)).....---------------- ( -14.50, z-score = -0.48, R) >droMoj3.scaffold_6500 21938471 99 + 32352404 -AGAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGCUGUCAUUUAAGUGCGG------GGAAUGCAAAGGAAAAAAAAAA-ACAAAAAAACAAUGAAACUGUAGAAA-- -...(((..(((.((((.(((((....(((....)))....))))).)))).------))).)))...............-..........................-- ( -16.20, z-score = -0.02, R) >droGri2.scaffold_15252 14367424 89 + 17193109 -AAAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGCUGUCAUUUUAGUGCGA------AGAAUGCGAAAGAAAAAAUAUUCACAUAUAAAAAAAGA------------- -...(((.....((((((((..((....))...))))).))).....)))((------(.....(....)........)))...............------------- ( -16.32, z-score = -0.73, R) >consensus AAAAGCAAAUCUGUGCGGUUAAACGCAAGUGUCAGCUGUCAUUUUUGUGGAA______AGAAUUCCGGC___AAAAAAUGCCAAGGAGCAUGAAAAAA__GCGCAGU__ ....(((((..((.((((((..((....))...))))))))..)))))............................................................. (-12.00 = -12.23 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:49:00 2011