| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,892,057 – 17,892,163 |
| Length | 106 |
| Max. P | 0.589960 |

| Location | 17,892,057 – 17,892,163 |
|---|---|
| Length | 106 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.76 |
| Shannon entropy | 0.40992 |
| G+C content | 0.33970 |
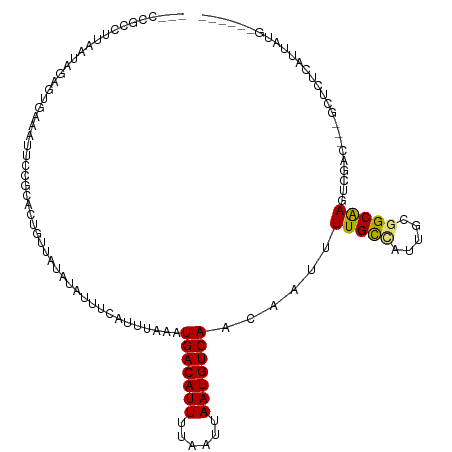
| Mean single sequence MFE | -19.98 |
| Consensus MFE | -10.81 |
| Energy contribution | -10.54 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.589960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17892057 106 - 23011544 ---CCGCCUUAAUAGAGUGAAAUUCCGCACUGUUAUAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAACAAUUUUGCCAUUGUGGCGAGUCGAC---GCUCUCAUUAUG------ ---..(((.((((((.(((......))).))))))...............(((((((......)))))))(((((......))))))))((((....---))))........------ ( -22.80, z-score = -2.20, R) >droAna3.scaffold_12916 13181401 106 + 16180835 ---CCGCCUUAAUAGACUGAAAUUCCGCACUGUUAUAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAACAAUUUUGCCAUUGCGGCAAGUCGAC---GCUCUCAUUAUG------ ---...........(((((((((...............))))))......(((((((......)))))))......(((((.....))))))))...---............------ ( -17.26, z-score = -0.95, R) >droEre2.scaffold_4845 16218814 109 + 22589142 CCACCGCCUUAAUAGAGUGAAAUUCCGCACUGUUAUAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAACAAUUUUGCCAUUGUGGCGAGUCGAC---GCUCUCAUUAUG------ .....(((.((((((.(((......))).))))))...............(((((((......)))))))(((((......))))))))((((....---))))........------ ( -22.80, z-score = -2.28, R) >droYak2.chr2R 17929971 106 + 21139217 ---CCGCCUUAAUAGAGUGAAAUUCCGCACUGUUAUAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAACAAUUUUGCCAUUGUGGCGAGUCGAC---GCUCUCAUUAUG------ ---..(((.((((((.(((......))).))))))...............(((((((......)))))))(((((......))))))))((((....---))))........------ ( -22.80, z-score = -2.20, R) >droSec1.super_7 1530572 106 - 3727775 ---CCGCCUUAAUAGAGUGAAAUUCCGCACUGUUAUAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAACAAUUUUGCCAUUGUGGCGAGUCGAC---GCUCUCAUUAUG------ ---..(((.((((((.(((......))).))))))...............(((((((......)))))))(((((......))))))))((((....---))))........------ ( -22.80, z-score = -2.20, R) >droSim1.chr2R 11961106 106 - 19596830 ---CCGCCUUAAUAGAGUGAAAUUCCGCACUGUUAUAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAACAAUUUUGCCAUUGUGGCGAGUCGAC---GCUCUCAUUAUG------ ---..(((.((((((.(((......))).))))))...............(((((((......)))))))(((((......))))))))((((....---))))........------ ( -22.80, z-score = -2.20, R) >droPer1.super_8 2246700 106 + 3966273 ---CUGCCUUAAUAGAGUGAAAUUCCUCACUGUUGUAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAACAAUUUUGCUAUUGCCGCAAGUCGAC---GCACUCAUUAUG------ ---.(((........(((((......)))))((((...............(((((((......))))))).....(((((.......))))).))))---))).........------ ( -18.90, z-score = -1.49, R) >dp4.chr4_group3 1186000 106 + 11692001 ---CUGCCUUAAUAGAGUGAAAUUCCUCACUGUUGUAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAACAAUUUUGCUAUUGCCGCAAGUCGAC---GCACUCAUUAUG------ ---.(((........(((((......)))))((((...............(((((((......))))))).....(((((.......))))).))))---))).........------ ( -18.90, z-score = -1.49, R) >droVir3.scaffold_12963 16933663 99 - 20206255 ----------------UUGUGUUGCCGAGCUGU--UAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAAC-AUUUUGUUAUUGCGGCAACUUUCUCAUCUGCUUAUUAAGUGUAUG ----------------....(((((((......--................((((((......))))))(((-.....)))....))))))).....(((..(((.....)))..))) ( -18.50, z-score = -1.23, R) >droMoj3.scaffold_6500 21934305 94 - 32352404 ------------------CUGCAGCCACGCUGU--UAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAAC-AUUUUGUUACUGCCGCAACUUUCU---CGUCUUAUUAGGCGUAUG ------------------..(((((...)))))--...............(((((((......)))))))..-........................---(((((....))))).... ( -15.40, z-score = -0.89, R) >droGri2.scaffold_15252 14364178 94 - 17193109 ------------------UUGUCGCGGCCCUGU--UAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAAC-AUUUUGUUAUUGCGGCAACUUUUC---CUUGUUAUUAGGCAUAUG ------------------(((((((((......--................((((((......))))))(((-.....))).))))))))).....(---((.......)))...... ( -16.80, z-score = -1.04, R) >consensus ___CCGCCUUAAUAGAGUGAAAUUCCGCACUGUUAUAUAUUUCAUUUAAAUGACAUUUUAAUUAAUGUCAACAAUUUUGCCAUUGCGGCAAGUCGAC___GCUCUCAUUAUG______ ..................................................(((((((......)))))))......(((((.....)))))........................... (-10.81 = -10.54 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:59 2011