
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,888,608 – 17,888,728 |
| Length | 120 |
| Max. P | 0.841026 |

| Location | 17,888,608 – 17,888,728 |
|---|---|
| Length | 120 |
| Sequences | 11 |
| Columns | 137 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Shannon entropy | 0.36696 |
| G+C content | 0.46463 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -15.05 |
| Energy contribution | -15.66 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.651267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
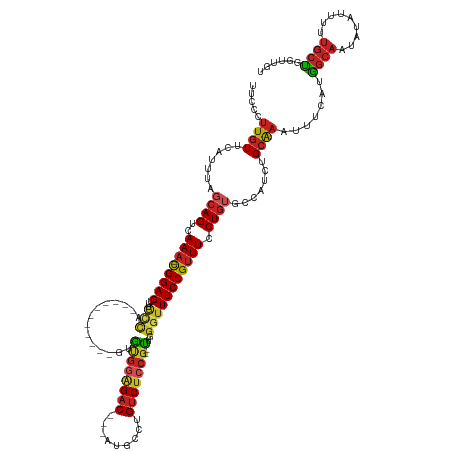
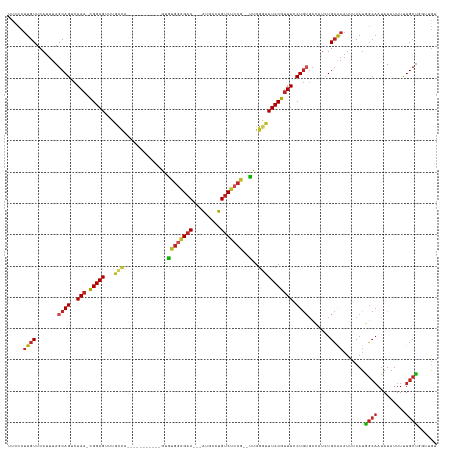
>dm3.chr2L 17888608 120 + 23011544 UUCCCUUGUUCAUUUAGCAGUCAAA-CGGAGUUUGCCA-----------GUGUGGAGAC---AUGCCUGUCUCCG--CUGGGUUUCCGUUUCCUGUGCCAUCUGCAAAUUUCAUGGCAAUAUAUUUUUGCUGGUUGU ..........((.(((((((..(((-(((((...(((.-----------..((((((((---(....))))))))--)..)))))))))))....((((((...........))))))........))))))).)). ( -39.30, z-score = -3.06, R) >droSim1.chr2L 17557401 120 + 22036055 UUCCCUUGUUCAUUUAGCAGUCAAA-CGGAGUUUGCCA-----------GUGUGGAGAC---AUGCCUGUCUCCG--CUGGGUUUCCGUUUCCUGUGCCAUCUGCAAAUUUCAUGGCAAUAUAUUUUUGCUGGUUGG ..........((.(((((((..(((-(((((...(((.-----------..((((((((---(....))))))))--)..)))))))))))....((((((...........))))))........))))))).)). ( -40.00, z-score = -3.02, R) >droSec1.super_7 1527093 120 + 3727775 UUCCCUUGUUCAUUUAGCAGUCAAA-CGGAGUUUGCCA-----------GUGUGGAGAC---AUCCCUGUCUCCG--CUGGGUUUCCGUUUCCUGUGCCAUCUGCAAAUUUCAUGGCAAUAUAUUUUUGCUGGUUGU ..........((.(((((((..(((-(((((...(((.-----------..((((((((---(....))))))))--)..)))))))))))....((((((...........))))))........))))))).)). ( -39.30, z-score = -3.51, R) >droYak2.chr2R 17926491 120 - 21139217 UUCCCUUGUUCAUUUAGCAGUCAAA-CGGAGUUUGCCA-----------GUGUGGAGAC---AUGCCUGUCUCCG--UUGGGUUUCCGUUUCCUGUGCCAUCUGCAAAUUUCAUGGCAAUAUAUUUUUGCUGGUUGU ..........((.(((((((..(((-(((((...(((.-----------...(((((((---(....))))))))--...)))))))))))....((((((...........))))))........))))))).)). ( -35.00, z-score = -2.01, R) >droEre2.scaffold_4845 16215430 120 - 22589142 UUCCCUUGUUCAUUUAGCAGUCAAA-CGGAGUUUGCCA-----------GUGUGGAGAC---AUGCCUGUCUCCG--CUGGGUUUCCGUUUCCUGUGCCAUCUGCAAAUUUCAUGGCAAUAUAUUUUUGCUGGUUGU ..........((.(((((((..(((-(((((...(((.-----------..((((((((---(....))))))))--)..)))))))))))....((((((...........))))))........))))))).)). ( -39.30, z-score = -3.06, R) >droAna3.scaffold_12916 13177848 120 - 16180835 UUUCCUUGUUCAUUUAGCAGUCAAA-CGGAGUUUGCCA-----------GUGUGGAGAC---AUGCCUGUCUCCG--CUGGGUUUCCGCUUCCUGUGCCAUCUGCAAAUUUCAUGGCAAUAUAUUUUUGCUGCUUGU ...............(((((.((((-(((((...(((.-----------..((((((((---(....))))))))--)..)))))))).......((((((...........)))))).......)))))))))... ( -38.40, z-score = -2.72, R) >dp4.chr4_group3 1182622 120 - 11692001 UUUCCUUGUUCAUUUAGCAGUCAAA-CGGAGUUUGCCA-----------GCAUGGAGAC---AUGCCUGUCUCCG--UCGGGUUUCCGUUUCCUGUGCCAUCUGCAAAUUUCAUGGCAAUAUAUUUCUGCUUCUGGU .........(((...(((((..(((-(((((...(((.-----------(.((((((((---(....))))))))--)).)))))))))))....((((((...........))))))........)))))..))). ( -37.10, z-score = -2.92, R) >droPer1.super_8 2243385 120 - 3966273 UUUCCUUGUUCAUUUAGCAGUCAAA-CGGAGUUUGCCA-----------GCAUGGAGAC---AUGCCUGUCUCCG--UCGGGUUUCCGUUUCCUGUGCCAUCUGCAAAUUUCAUGGCAAUAUAUUUCUGCUUCUGGU .........(((...(((((..(((-(((((...(((.-----------(.((((((((---(....))))))))--)).)))))))))))....((((((...........))))))........)))))..))). ( -37.10, z-score = -2.92, R) >droWil1.scaffold_180764 3423004 122 - 3949147 UUGCCUUGUUCACUUAGCAGUCAAA-UGGAGGCUGUUUCUCUC---UCUGAGCGGAGAC---AUAGCUGUCUCCG--UUGGGUUUCCGUUUCCUAUGGUCUCUGGG----CCAUAU--AUAUAUUUCUGCUUACUGA .........(((...(((((..(((-((((((((....(((..---...)))(((((((---(....))))))))--...)))))))))))..(((((((....))----))))).--........)))))...))) ( -41.20, z-score = -3.24, R) >droMoj3.scaffold_6500 21928063 126 + 32352404 UUGGCUUGUUCAUUUAGCAGUCAAAGCGGAGUUGCCUUCGAA-------GGACAAUGCC--CGGCGCUGUCUCCGCGUUGGCUUUCCGUUUCCUG--CCUCGUGCAAAUUUCAUGGCAAUAUAUUUUUGCCGGCCAA .((((((((.......((((...((((((((..(((..((..-------(((((.(((.--..))).)))))...))..))).)))))))).)))--).....))))......((((((.......)))))))))). ( -42.70, z-score = -1.75, R) >droGri2.scaffold_15252 14359722 132 + 17193109 UUGCCUUGUUCACUUAGCAGCCAAA-CGGAGUCGACGACGACAAAGACUGGACGAGGACAACGAUGCUGUCUCCA--UUGGCUUUCCGUUUCCUG--CCUUGUGCAAAUUUCAUGGCAAUAUAUUUUUGCUGGCCAA ..((((((..(((...((((..(((-((((((((....)))).(((.(..(..(..((((.......))))..).--)..))))))))))).)))--)...)))))).......(((((.......))))))))... ( -34.40, z-score = -0.26, R) >consensus UUCCCUUGUUCAUUUAGCAGUCAAA_CGGAGUUUGCCA___________GUGUGGAGAC___AUGCCUGUCUCCG__UUGGGUUUCCGUUUCCUGUGCCAUCUGCAAAUUUCAUGGCAAUAUAUUUUUGCUGGUUGU ................((((.......((((......................((((((.........))))))....(((....))).))))........)))).........((((.........))))...... (-15.05 = -15.66 + 0.61)

| Location | 17,888,608 – 17,888,728 |
|---|---|
| Length | 120 |
| Sequences | 11 |
| Columns | 137 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Shannon entropy | 0.36696 |
| G+C content | 0.46463 |
| Mean single sequence MFE | -35.65 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.71 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.841026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
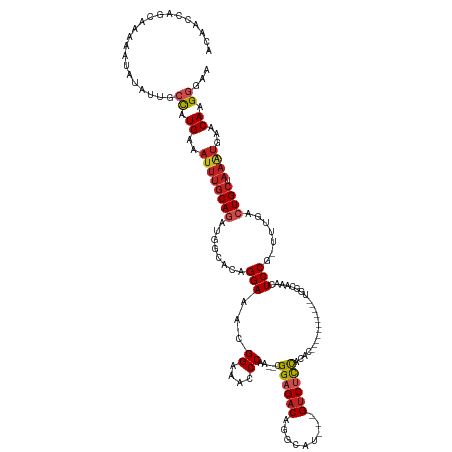
>dm3.chr2L 17888608 120 - 23011544 ACAACCAGCAAAAAUAUAUUGCCAUGAAAUUUGCAGAUGGCACAGGAAACGGAAACCCAG--CGGAGACAGGCAU---GUCUCCACAC-----------UGGCAAACUCCG-UUUGACUGCUAAAUGAACAAGGGAA ....((.............((((((...........))))))(((.(((((((...((((--.(((((((....)---))))))...)-----------))).....))))-)))..)))............))... ( -35.10, z-score = -2.71, R) >droSim1.chr2L 17557401 120 - 22036055 CCAACCAGCAAAAAUAUAUUGCCAUGAAAUUUGCAGAUGGCACAGGAAACGGAAACCCAG--CGGAGACAGGCAU---GUCUCCACAC-----------UGGCAAACUCCG-UUUGACUGCUAAAUGAACAAGGGAA ....((.............((((((...........))))))(((.(((((((...((((--.(((((((....)---))))))...)-----------))).....))))-)))..)))............))... ( -35.10, z-score = -2.57, R) >droSec1.super_7 1527093 120 - 3727775 ACAACCAGCAAAAAUAUAUUGCCAUGAAAUUUGCAGAUGGCACAGGAAACGGAAACCCAG--CGGAGACAGGGAU---GUCUCCACAC-----------UGGCAAACUCCG-UUUGACUGCUAAAUGAACAAGGGAA ....((.............((((((...........))))))(((.(((((((...((((--.(((((((....)---))))))...)-----------))).....))))-)))..)))............))... ( -35.10, z-score = -2.69, R) >droYak2.chr2R 17926491 120 + 21139217 ACAACCAGCAAAAAUAUAUUGCCAUGAAAUUUGCAGAUGGCACAGGAAACGGAAACCCAA--CGGAGACAGGCAU---GUCUCCACAC-----------UGGCAAACUCCG-UUUGACUGCUAAAUGAACAAGGGAA ....((.............((((((...........))))))(((.(((((((...(((.--.(((((((....)---))))))....-----------))).....))))-)))..)))............))... ( -32.00, z-score = -2.21, R) >droEre2.scaffold_4845 16215430 120 + 22589142 ACAACCAGCAAAAAUAUAUUGCCAUGAAAUUUGCAGAUGGCACAGGAAACGGAAACCCAG--CGGAGACAGGCAU---GUCUCCACAC-----------UGGCAAACUCCG-UUUGACUGCUAAAUGAACAAGGGAA ....((.............((((((...........))))))(((.(((((((...((((--.(((((((....)---))))))...)-----------))).....))))-)))..)))............))... ( -35.10, z-score = -2.71, R) >droAna3.scaffold_12916 13177848 120 + 16180835 ACAAGCAGCAAAAAUAUAUUGCCAUGAAAUUUGCAGAUGGCACAGGAAGCGGAAACCCAG--CGGAGACAGGCAU---GUCUCCACAC-----------UGGCAAACUCCG-UUUGACUGCUAAAUGAACAAGGAAA ...((((((..........((((((...........))))))....(((((((...((((--.(((((((....)---))))))...)-----------))).....))))-)))).)))))............... ( -36.70, z-score = -2.86, R) >dp4.chr4_group3 1182622 120 + 11692001 ACCAGAAGCAGAAAUAUAUUGCCAUGAAAUUUGCAGAUGGCACAGGAAACGGAAACCCGA--CGGAGACAGGCAU---GUCUCCAUGC-----------UGGCAAACUCCG-UUUGACUGCUAAAUGAACAAGGAAA ..((..(((((........((((((...........))))))....(((((((..((.(.--.(((((((....)---))))))...)-----------.)).....))))-)))..)))))...)).......... ( -33.90, z-score = -2.45, R) >droPer1.super_8 2243385 120 + 3966273 ACCAGAAGCAGAAAUAUAUUGCCAUGAAAUUUGCAGAUGGCACAGGAAACGGAAACCCGA--CGGAGACAGGCAU---GUCUCCAUGC-----------UGGCAAACUCCG-UUUGACUGCUAAAUGAACAAGGAAA ..((..(((((........((((((...........))))))....(((((((..((.(.--.(((((((....)---))))))...)-----------.)).....))))-)))..)))))...)).......... ( -33.90, z-score = -2.45, R) >droWil1.scaffold_180764 3423004 122 + 3949147 UCAGUAAGCAGAAAUAUAU--AUAUGG----CCCAGAGACCAUAGGAAACGGAAACCCAA--CGGAGACAGCUAU---GUCUCCGCUCAGA---GAGAGAAACAGCCUCCA-UUUGACUGCUAAGUGAACAAGGCAA (((.(.(((((........--.(((((----........)))))(....)((....))..--((((((((....)---))))))).(((((---..(((.......)))..-)))))))))).).)))......... ( -35.00, z-score = -3.50, R) >droMoj3.scaffold_6500 21928063 126 - 32352404 UUGGCCGGCAAAAAUAUAUUGCCAUGAAAUUUGCACGAGG--CAGGAAACGGAAAGCCAACGCGGAGACAGCGCCG--GGCAUUGUCC-------UUCGAAGGCAACUCCGCUUUGACUGCUAAAUGAACAAGCCAA .((((.(((((.......))))).((..((((((((((((--(.(....)(((..(((..((((....).)))...--))).(((.((-------(....)))))).)))))))))..))).))))...)).)))). ( -42.00, z-score = -2.01, R) >droGri2.scaffold_15252 14359722 132 - 17193109 UUGGCCAGCAAAAAUAUAUUGCCAUGAAAUUUGCACAAGG--CAGGAAACGGAAAGCCAA--UGGAGACAGCAUCGUUGUCCUCGUCCAGUCUUUGUCGUCGUCGACUCCG-UUUGGCUGCUAAGUGAACAAGGCAA ...(((.((((.......))))...........(((..((--(((.(((((((.......--((((((((((...))))))....))))......((((....))))))))-)))..)))))..))).....))).. ( -38.20, z-score = -0.77, R) >consensus ACAACCAGCAAAAAUAUAUUGCCAUGAAAUUUGCAGAUGGCACAGGAAACGGAAACCCAA__CGGAGACAGGCAU___GUCUCCACAC___________UGGCAAACUCCG_UUUGACUGCUAAAUGAACAAGGGAA .....................((.((..((((((((........(((...((....)).....((((((.........)))))).......................))).......)))).))))...)).))... (-17.00 = -17.71 + 0.71)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:58 2011