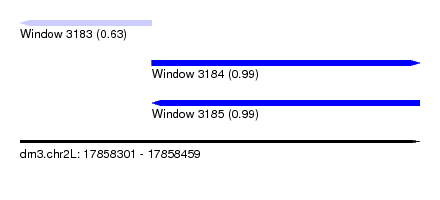
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,858,301 – 17,858,459 |
| Length | 158 |
| Max. P | 0.992317 |

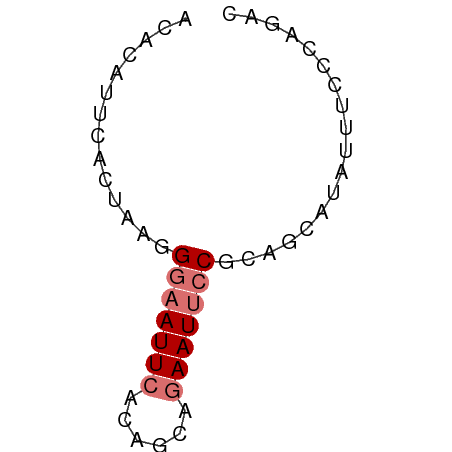
| Location | 17,858,301 – 17,858,353 |
|---|---|
| Length | 52 |
| Sequences | 4 |
| Columns | 52 |
| Reading direction | reverse |
| Mean pairwise identity | 85.58 |
| Shannon entropy | 0.23402 |
| G+C content | 0.42996 |
| Mean single sequence MFE | -7.98 |
| Consensus MFE | -4.70 |
| Energy contribution | -5.45 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17858301 52 - 23011544 ACACAUUCACUAAGGAAAUUCACAGCAGAAUUCCGCAUCAUAUUUCCCAGAC .............((((((.....((.(....).)).....))))))..... ( -6.60, z-score = -2.90, R) >droSim1.chr2L 17536473 52 - 22036055 ACACAUUCACUAAGGGAAUUCACAGCAGAAUUCCGCAGCAUAUUUCCCAGAC ..............(((((((......))))))).................. ( -8.80, z-score = -3.06, R) >droSec1.super_7 1491212 52 - 3727775 ACACAUUCACUAAGGGAAUUCACAGCAGAAUUCCGCAGCAUAUUUCCCAGAC ..............(((((((......))))))).................. ( -8.80, z-score = -3.06, R) >droEre2.scaffold_4845 16185231 51 + 22589142 -CAAAUCCACUAAGGGAAUUUAAAGCCUAAUGCCUCAGCAUUUCUCUCGGAC -........(..((((((...........((((....))))))))))..).. ( -7.70, z-score = -0.42, R) >consensus ACACAUUCACUAAGGGAAUUCACAGCAGAAUUCCGCAGCAUAUUUCCCAGAC ..............(((((((......))))))).................. ( -4.70 = -5.45 + 0.75)

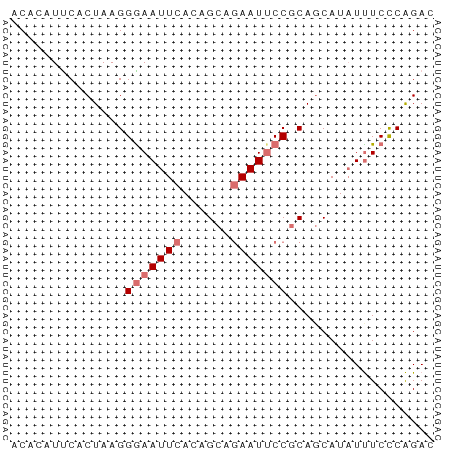
| Location | 17,858,353 – 17,858,459 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 94.34 |
| Shannon entropy | 0.07797 |
| G+C content | 0.31447 |
| Mean single sequence MFE | -20.33 |
| Consensus MFE | -18.59 |
| Energy contribution | -18.27 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.992317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17858353 106 + 23011544 UAAAUACCAAGAGAUAUGGGCUGUUUCGAAAUUAACCAGCAUUGUAGCCAAUUACUGAAAUUUUACUUUUGUAAUUAGUCCUACUAUUCCUAUCUAUAUUUCAUAG ..........(((((((((((((((........)).))))...((((.(((((((.(((.......))).)))))).)..)))).........))))))))).... ( -18.70, z-score = -1.62, R) >droSim1.chr2L 17536525 106 + 22036055 UAAAUACCAUGAGAUAUGGACUGUUUCGAAAUUAACCAGCAUUGUAGCCGAUUACUGAAAUUUUACUUUCGUAAUUACUCCUACUAUUCCAAACCAUAUUUCAUAG ........(((((((((((.(((((........)).)))....((((..((((((.((((......))))))))))....)))).........))))))))))).. ( -21.10, z-score = -3.75, R) >droSec1.super_7 1491264 106 + 3727775 UAAAUACCAUGAGAUAUGGACUGUUUCGAAAUUAACCAGCAUUGUAGCCGAUUACUGAAAUUUUACUUUCGUAAUUACUCCUACUAAUCCUAACCAUAUUUCAUAG ........(((((((((((...(((........))).((.(((((((..((((((.((((......))))))))))....)))).))).))..))))))))))).. ( -21.20, z-score = -3.79, R) >consensus UAAAUACCAUGAGAUAUGGACUGUUUCGAAAUUAACCAGCAUUGUAGCCGAUUACUGAAAUUUUACUUUCGUAAUUACUCCUACUAUUCCUAACCAUAUUUCAUAG ........(((((((((((.(((((........)).)))....((((..((((((.((((......))))))))))....)))).........))))))))))).. (-18.59 = -18.27 + -0.33)

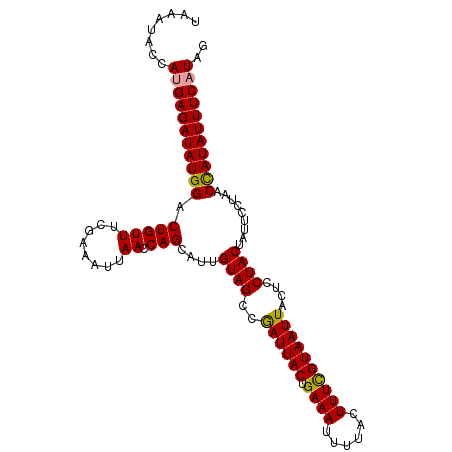
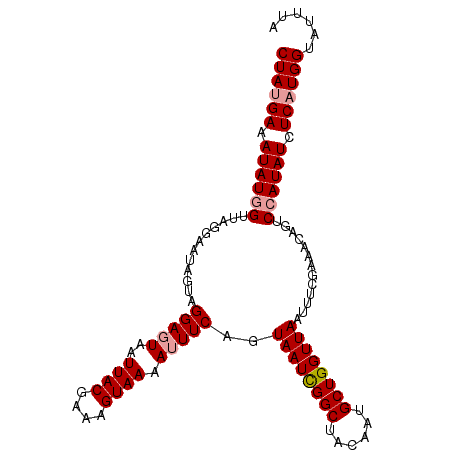
| Location | 17,858,353 – 17,858,459 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 94.34 |
| Shannon entropy | 0.07797 |
| G+C content | 0.31447 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -22.29 |
| Energy contribution | -23.07 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17858353 106 - 23011544 CUAUGAAAUAUAGAUAGGAAUAGUAGGACUAAUUACAAAAGUAAAAUUUCAGUAAUUGGCUACAAUGCUGGUUAAUUUCGAAACAGCCCAUAUCUCUUGGUAUUUA .....(((((((((((((....((((..((((((((..((((...))))..))))))))))))...((((.............)))))).)))))....)))))). ( -20.22, z-score = -0.98, R) >droSim1.chr2L 17536525 106 - 22036055 CUAUGAAAUAUGGUUUGGAAUAGUAGGAGUAAUUACGAAAGUAAAAUUUCAGUAAUCGGCUACAAUGCUGGUUAAUUUCGAAACAGUCCAUAUCUCAUGGUAUUUA ((((((.((((((((((((((....(((((..((((....)))).)))))..((((((((......)))))))))))))))).....)))))).))))))...... ( -32.20, z-score = -5.05, R) >droSec1.super_7 1491264 106 - 3727775 CUAUGAAAUAUGGUUAGGAUUAGUAGGAGUAAUUACGAAAGUAAAAUUUCAGUAAUCGGCUACAAUGCUGGUUAAUUUCGAAACAGUCCAUAUCUCAUGGUAUUUA ((((((.((((((....(.......(((((..((((....)))).)))))..((((((((......)))))))).........)...)))))).))))))...... ( -28.50, z-score = -3.79, R) >consensus CUAUGAAAUAUGGUUAGGAAUAGUAGGAGUAAUUACGAAAGUAAAAUUUCAGUAAUCGGCUACAAUGCUGGUUAAUUUCGAAACAGUCCAUAUCUCAUGGUAUUUA ((((((.((((((............(((((..((((....)))).)))))..((((((((......)))))))).............)))))).))))))...... (-22.29 = -23.07 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:51 2011