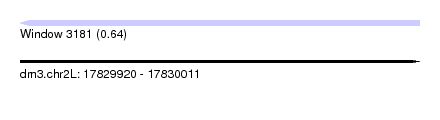
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,829,920 – 17,830,011 |
| Length | 91 |
| Max. P | 0.643870 |

| Location | 17,829,920 – 17,830,011 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.05 |
| Shannon entropy | 0.35072 |
| G+C content | 0.44682 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -12.28 |
| Energy contribution | -13.67 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17829920 91 - 23011544 UCGGGCAAGUUUAUAUUCAUUAAACUUCCGCCACAUUUUGCGCUUUAUAUUUUGGCGCUUUC----AAAUGUCGGCCCGGGACACUC----GACUGAAG---- (((((((((((((.......))))))).((..((((((.(((((.........)))))....----)))))))))))))).......----........---- ( -25.00, z-score = -2.38, R) >droSim1.chr2L 17507416 95 - 22036055 UCGGGCAAGUUUAUAUUCAUUAAACUUCCGCCACAUUUUGCGCUUUAUAUUUUGGCGCUUUC----AAAUGUCGGCCCGGGACACUC----ACUCGACUGAAG ...((.(((((((.......)))))))))(((((((((.(((((.........)))))....----)))))).))).((((......----.))))....... ( -25.40, z-score = -2.56, R) >droSec1.super_7 1462768 95 - 3727775 UCGGGCAAGUUUAUAUUCAUUAAACUUCCGCCACAUUUUGCGCUUUAUAUUUUGGCGCUUUC----AAAUGUCGGCCCGGGACACUC----ACUCGACUGAAG ...((.(((((((.......)))))))))(((((((((.(((((.........)))))....----)))))).))).((((......----.))))....... ( -25.40, z-score = -2.56, R) >droYak2.chr2R 17864317 91 + 21139217 UCGGGCAAGUUUAUAUUCAUUAAACUUCCGCCACAUUUUGCGCUUUAUAUUUUGGCGCUUUC----AAAUGUCGGCCCGGGACACUC----GACUGAAG---- (((((((((((((.......))))))).((..((((((.(((((.........)))))....----)))))))))))))).......----........---- ( -25.00, z-score = -2.38, R) >droEre2.scaffold_4845 16157129 91 + 22589142 UCGGGCAAGUUUAUAUUCAUUAAACUUCCGCCACAUUUUGCGCUUUAUAUUUUGGCGCUUUC----AAAUGUCGGCCCGGGACUCUC----GACUGACG---- (((((((((((((.......))))))).((..((((((.(((((.........)))))....----)))))))))))))).......----........---- ( -25.00, z-score = -2.44, R) >droAna3.scaffold_12916 13119323 83 + 16180835 UCGGCCAAGUUUAUAUUCAUUAAACUUCCGCCACAUUUUGCGCUUUAUAUUUUGGCGCUUUC----AAAUGUCGGCCCGGGACACUC---------------- ((((..(((((((.......)))))))..(((((((((.(((((.........)))))....----)))))).))))))).......---------------- ( -22.70, z-score = -2.81, R) >dp4.chr4_group3 1122710 90 + 11692001 UCGAGCAAGUUUAUAUUCAUUAAACUUCCGCCACAUUUUGCGCUUUAUAUUUUGGCGCUUUC----AAAUGUC----GGUGGCCGCC----ACUCGAGGCUC- (((((.(((((((.......)))))))(((((((((((.(((((.........)))))....----)))))).----))))).....----.))))).....- ( -28.20, z-score = -3.32, R) >droPer1.super_8 2181074 94 + 3966273 UCGAGCAAGUUUAUAUUCAUUAAACUUCCGCCACAUUUUGCGCUUUAUAUUUUGGCGCUUUC----AAAUGUCGGGGGGGGGCCGCC----ACUCGAGGCUC- ..(((((((((((.......)))))))((.((((((((.(((((.........)))))....----)))))).)).))(((......----.)))...))))- ( -27.70, z-score = -1.41, R) >droWil1.scaffold_180764 3321440 102 + 3949147 UCAGACAAGUUUAUAUUCAUUAAACUUCCGCCAAAUUUUGCGCCUUAUAUUUUAGACAUUUCACCAAAUUGCCGCACUGGGACUUUCACUAACUCUAUGUUC- ((((..(((((((.......)))))))...........((((.(..........................).))))))))......................- ( -8.57, z-score = 1.07, R) >consensus UCGGGCAAGUUUAUAUUCAUUAAACUUCCGCCACAUUUUGCGCUUUAUAUUUUGGCGCUUUC____AAAUGUCGGCCCGGGACACUC____ACUCGACG____ ((((....(((((.......)))))(((((((((((((.(((((.........)))))........)))))).))..)))))...........))))...... (-12.28 = -13.67 + 1.38)

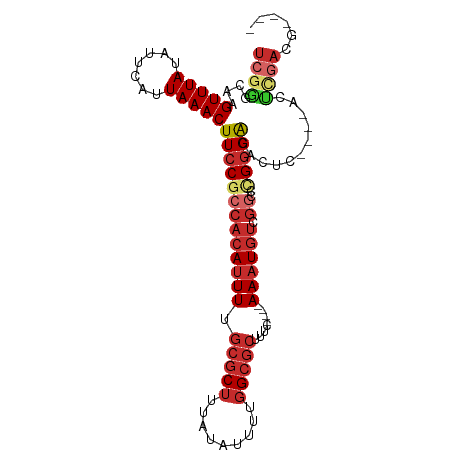
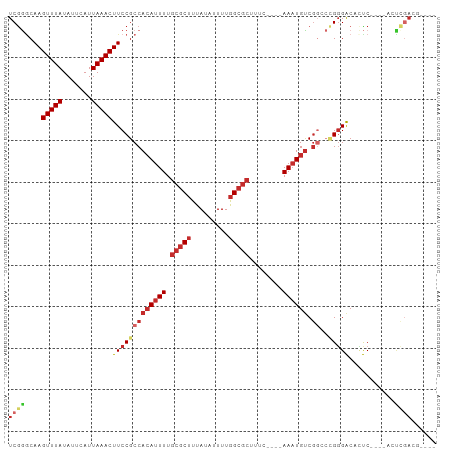
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:48 2011