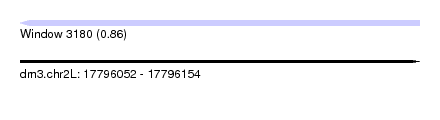
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,796,052 – 17,796,154 |
| Length | 102 |
| Max. P | 0.857304 |

| Location | 17,796,052 – 17,796,154 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.14 |
| Shannon entropy | 0.67952 |
| G+C content | 0.29421 |
| Mean single sequence MFE | -15.32 |
| Consensus MFE | -6.21 |
| Energy contribution | -6.74 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17796052 102 - 23011544 ---UUUUAGUCAUGUCUUCUACGGCAUA-----------AGUUCAAUAUUUUGUAGAAGUU----CUUUCUAUCAAAGUAUCCUAGUUCCUAAGCUUAAAAAAUCUUGCCAUCAUGUUCA ---...................((((..-----------......((((((((((((((..----.)))))).))))))))...(((......)))..........)))).......... ( -13.80, z-score = -1.56, R) >droSim1.chr2L 17478233 105 - 22036055 CAUGUUUCUUUAUGUCUUCUACGGAAUA-----------AGUUCAAUAUUUUGUAGAAGAC----CUUUCUAUCAAAGUAUCCAAGUUCCUAAGCUUAAAAAAUCUUUCCAUCAUGUUCA ((((.........(((((((((((((((-----------.......)))))))))))))))----..................((((......))))...............)))).... ( -20.50, z-score = -3.74, R) >droSec1.super_7 1430745 102 - 3727775 ---GUUUUAUUGUGUCUUCUACGGAAUA-----------AGUUCAAUAUUUUGUAGAAGUC----CUUUCUAUCAAAGUAUCCAAGUUCCUAAGCUUAAAAAAUCUUUCCAUCAUGUUCA ---..........(.(((((((((((((-----------.......))))))))))))).)----..................((((......))))....................... ( -14.60, z-score = -2.30, R) >droYak2.chr2R 17835851 115 + 21139217 ---AUUUUUUUUUAUCUUCUAGGACAUGACAAUUGUAUCAAUGCUAUAUUUUUUAGAAGUCUAAACUUUUGAUUGAAGU-UCCUAAUUCCUAAGCUU-AAAAUUCGUUGCGUCGUGUUCA ---..................(((((((((..................((..((((((((....))))))))..))...-.............((..-..........))))))))))). ( -19.50, z-score = -1.36, R) >droEre2.scaffold_4845 16128934 113 + 22589142 ---ACUUUAAUAUAUAUUCUAGGGAAUA--AAAUUGAUCAAUAUUAUAUUUUUUAGAAGUCCCUAAUUUUCGUUAAAGU-UCCCAAGUCCUAGGCUU-AAAAUUCCUUUCGUCGUGUUCA ---((((((((........((((((.((--(((.((((....))))....)))))....))))))......))))))))-....((((.....))))-...................... ( -14.54, z-score = -0.22, R) >droVir3.scaffold_13269 6280 88 - 8408 ---GGUUCAUUAAACAAUUUAUUGAAUG---------CGUAAGUAAUCUUUUAUAUUAA------CCUACUACUAGAA----CGAGUACUUGAGA---AAAAGUCUAUAUGUA------- ---.((((......(((....)))..((---------(....)))..............------..........)))----).((.((((....---..)))))).......------- ( -9.00, z-score = 1.14, R) >consensus ___GUUUUAUUAUAUCUUCUACGGAAUA___________AAUUCAAUAUUUUGUAGAAGUC____CUUUCUAUCAAAGU_UCCAAGUUCCUAAGCUU_AAAAAUCUUUCCAUCAUGUUCA ...............(((((((((((((..................)))))))))))))............................................................. ( -6.21 = -6.74 + 0.53)

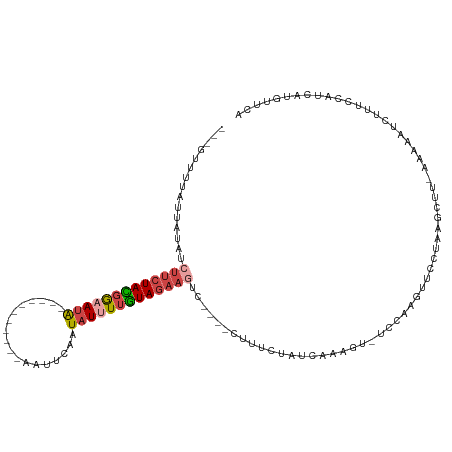
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:47 2011