| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,724,783 – 17,724,875 |
| Length | 92 |
| Max. P | 0.959302 |

| Location | 17,724,783 – 17,724,875 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 67.43 |
| Shannon entropy | 0.65906 |
| G+C content | 0.41734 |
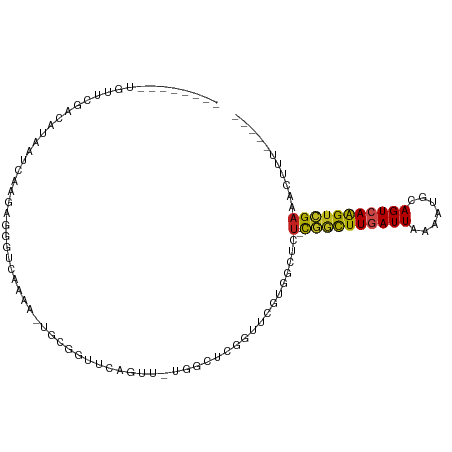
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -9.43 |
| Energy contribution | -9.07 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.90 |
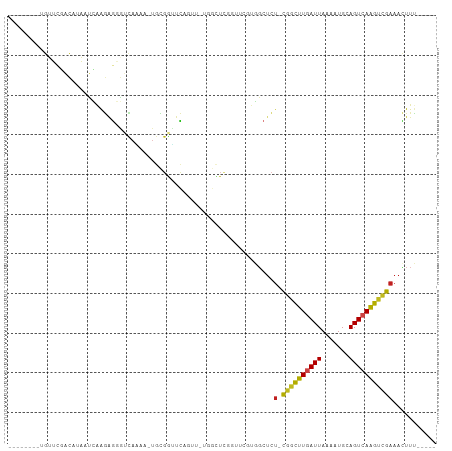
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
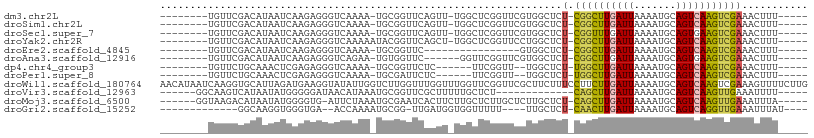
>dm3.chr2L 17724783 92 + 23011544 --------UGUUCGACAUAAUCAAGAGGGUCAAAA-UGCGGUUCAGUU-UGGCUCGGUUCGUGGCUCU-CGGCUUGAUUAAAAUGCAGUCAAGUCGAAACUUU----- --------..(((...........((((((((...-..(((..(....-.)..))).....)))))))-)(((((((((.......)))))))))))).....----- ( -25.10, z-score = -1.70, R) >droSim1.chr2L 17404936 92 + 22036055 --------UGUUCGACAUAAUCAAGAGGGUCAAAA-UGCGGUUCAGUU-UGGCUCGGUUCGUGGCUCU-CGGCUUGAUUAAAAUGCAGUCAAGUCGAAACUUU----- --------..(((...........((((((((...-..(((..(....-.)..))).....)))))))-)(((((((((.......)))))))))))).....----- ( -25.10, z-score = -1.70, R) >droSec1.super_7 1360821 92 + 3727775 --------UGUUCGACAUAAUCAAGAGGGUCAAAA-UGCGGUUCAGUU-UGGCUCGGUUCGUGGCUCU-CGGUUUGAUUAAAAUGCAGUGAAGUCGAAACUUU----- --------..((((((.(((((((((((((((...-..(((..(....-.)..))).....)))))))-)...)))))))...(.......))))))).....----- ( -21.90, z-score = -0.60, R) >droYak2.chr2R 17764832 93 - 21139217 --------UGUUCGACAUAAUCAAGAGGGUCAAAAAUACGGUUCAGCU-UGGCUCGGUUCCUGGCUCU-CGGCUUGAUUAAAAUGCAGUCAAGUCGAAACUUU----- --------..(((...........((((((((..(((.(((..(....-.)..))))))..)))))))-)(((((((((.......)))))))))))).....----- ( -25.00, z-score = -1.75, R) >droEre2.scaffold_4845 16056812 77 - 22589142 --------UGUUCGACAUAAUCAAGAGGGUCAAAA-UGCGGUUC----------------GUGGCUCU-CGGCUUGAUUAAAAUGCAGUCAAGUCGAAACUUU----- --------..(((...........((((((((..(-......).----------------.)))))))-)(((((((((.......)))))))))))).....----- ( -21.80, z-score = -2.46, R) >droAna3.scaffold_12916 13018145 87 - 16180835 --------UGUUCGACAUAAUCAAGAGGGUCAGAA-UGUGGUUC------GGUUCGGUUCGUGGCUCU-CGGCUUGAUUAAAAUGCAGUGAAGUCGAAACUUU----- --------..((((((........(((((((((((-(.(((...------...))))))).)))))))-).((...........))......)))))).....----- ( -23.00, z-score = -1.14, R) >dp4.chr4_group3 1003735 85 - 11692001 --------UGUUCUGCAAACUCGAGAGGGUCAAAA-UGCGGUUCUC------UUCGGUU--UGGCUCU-UGGCUUGAUUAAAAUGCAGUCAAGUCGAAACUUU----- --------.......((((((.(((((..((....-...))..)))------)).))))--))....(-((((((((((.......)))))))))))......----- ( -27.10, z-score = -2.93, R) >droPer1.super_8 2063494 85 - 3966273 --------UGUUCUGCAAACUCGAGAGGGUCAAAA-UGCGAUUCUC------UUCGGUU--UGGCUCU-UGGCUUGAUUAAAAUGCAGUCAAGUCGAAACUUU----- --------.......((((((.(((((((((....-...)))))))------)).))))--))....(-((((((((((.......)))))))))))......----- ( -28.30, z-score = -3.37, R) >droWil1.scaffold_180764 3203131 108 - 3949147 AACAUAAUCAAGGUGCAUUAGAUGAAGGUAUAUUGGUCUUGGUUUGGUUUGGUUCGGUUCGCUUCUUUCCUUCUUGAUUAAAAUGCAGUCAAGUCGAAAGUUUUCUUG ........(((((((.((..(((.(((.((.(((......))).)).))).)))..)).)))..(((((...(((((((.......)))))))..)))))....)))) ( -18.90, z-score = 0.40, R) >droVir3.scaffold_12963 16706483 84 + 20206255 ------GGCAAGUCAUAAUAUGGGGGAUAACAUAAAUGCGGUUCGCUUUUUGCUCU-------------CAGCUUGAUUAAAAUGCAGUCAAGUUGAAAUUUU----- ------((((((......((((........))))...(((...)))..)))))).(-------------((((((((((.......)))))))))))......----- ( -19.50, z-score = -1.49, R) >droMoj3.scaffold_6500 21663331 95 + 32352404 ------GGUAAGACAUAAUAUGGGGUG-AUUCUAAAUGCGAAUCACUUCUUGCUCUUGCUCUUGCUCU-CAGCUUGAUUAAAAUGCAGUCAAGUUGAAAUUUA----- ------(((((((((......((((((-((((.......)))))))))).......)).))))))).(-((((((((((.......)))))))))))......----- ( -31.12, z-score = -4.56, R) >droGri2.scaffold_15252 14121634 83 + 17193109 -------------GGCAAGGUGGGUGA--ACCAAAAUGCGG-UUGAUGGUGGUUUUU----UUGCUCU-CAACUUGAUUAAAAUGCAGUCAGGUUGAAAUUUAU---- -------------.....((..((.((--((((..((....-...))..)))))).)----)..)).(-((((((((((.......))))))))))).......---- ( -21.50, z-score = -1.50, R) >consensus ________UGUUCGACAUAAUCAAGAGGGUCAAAA_UGCGGUUCAGUU_UGGCUCGGUUCGUGGCUCU_CGGCUUGAUUAAAAUGCAGUCAAGUCGAAACUUU_____ .....................................................................((((((((((.......))))))))))............ ( -9.43 = -9.07 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:41 2011