
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,740,597 – 1,740,701 |
| Length | 104 |
| Max. P | 0.702995 |

| Location | 1,740,597 – 1,740,701 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.01 |
| Shannon entropy | 0.65157 |
| G+C content | 0.34768 |
| Mean single sequence MFE | -21.05 |
| Consensus MFE | -7.40 |
| Energy contribution | -7.25 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1740597 104 - 23011544 ---AUUAUUCCGCAGUUAGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUUCUGCCCGAUUUAGGGUUUGCUAAUUACUUAUGCAAACAAAU--------- ---........((.....))...(((((((................((((....))))((((((....((((......)))).....))))))....)))))))...--------- ( -23.80, z-score = -2.09, R) >droSim1.chr2L 1731757 104 - 22036055 ---AUUAUUCCGCAGUUCGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUUCUGCCCGAUUUAGGGUUUGCUAAUUACUUAUGCAAACAAAU--------- ---........((.....))...(((((((................((((....))))((((((....((((......)))).....))))))....)))))))...--------- ( -23.80, z-score = -2.33, R) >droSec1.super_14 1674720 104 - 2068291 ---AUUAUUCCGCAGUUCGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUUCUGCCCGAUUUAGGGUUUGCUAAUUACUUAUGCAAACAAAU--------- ---........((.....))...(((((((................((((....))))((((((....((((......)))).....))))))....)))))))...--------- ( -23.80, z-score = -2.33, R) >droYak2.chr2L 1711484 104 - 22324452 ---AUUAUUCUCCAGUUAGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUUCUGCCCGAUUUAGGGUUUGCUAAUUACUUAUGCAAACAAAU--------- ---....................(((((((................((((....))))((((((....((((......)))).....))))))....)))))))...--------- ( -23.00, z-score = -2.18, R) >droEre2.scaffold_4929 1784727 104 - 26641161 ---UCUAUUCCUCAGUUAGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUUUUGCCCGAUUUAGGGUUAGCUAAUUACUUAUGCAAACAAAU--------- ---....................(((((((................((((....))))((((((....((((......)))).....))))))....)))))))...--------- ( -23.00, z-score = -2.03, R) >droAna3.scaffold_12916 335282 95 + 16180835 AAAGUUCUGGUCAAAUUAGGCGCUAUUUGCUAAUUUCGUAUACCCUUGGCUUAGGUCAGU-------UUCUA-UUUGGCAGCUA---AAC-GACUAAGUAAACAAAU--------- .......((((((((((((.((.....))))))))).........((((((...(((((.-------.....-.))))))))))---)..-)))))...........--------- ( -16.30, z-score = 0.57, R) >dp4.chr4_group3 8463456 94 - 11692001 ---UUUGCUGUCCGAUUAGCUAAGCUCUGUUAAUUUGUCCAAACCUUGUCAUAGGUCAGG-------UUCGAGGCCUAUGUUCA---AACAAAUCAAAUAAACAAAU--------- ---...((((......)))).......((((.(((((........(((.((((((((...-------.....))))))))..))---)......))))).))))...--------- ( -20.04, z-score = -2.08, R) >droPer1.super_1 9914443 94 - 10282868 ---UUUGCCGUCCGAUUAGCUAAGCUCUGUUAAUUUGUCAAAACCUUGUCAUAGGUCAGG-------UUCGAGGCCUAUGUUCA---AACAAAUCAAAUAAACAAAU--------- ---((((..(.(.(((((((........))))))).).)......(((.((((((((...-------.....))))))))..))---).))))..............--------- ( -18.00, z-score = -1.54, R) >droWil1.scaffold_181132 185701 96 + 1035393 ------------GAAUUGACUAGAAUUUGCUAAUUUUAUUGCCCAUUGGGUUAGGUCAG--------UUCCUGUUUUUGGACGGUCUAACAAUUUAAGCAAACUAAGAUUUUAAGU ------------(((((((((((((((....))))))...((((...))))..))))))--------)))....((((((...(..(((....)))..)...))))))........ ( -17.70, z-score = -0.03, R) >consensus ___AUUAUUCCCCAGUUAGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUU_UGCCCGAUUUAGGGUUAGCUAAUUACUUAUGCAAACAAAU_________ .............(((((((........)))))))..............((((((((...............)))))))).................................... ( -7.40 = -7.25 + -0.16)
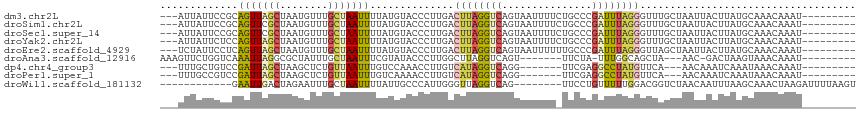

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:09:00 2011