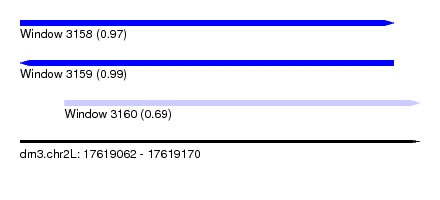
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,619,062 – 17,619,170 |
| Length | 108 |
| Max. P | 0.989812 |

| Location | 17,619,062 – 17,619,163 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.65 |
| Shannon entropy | 0.56897 |
| G+C content | 0.49419 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -14.19 |
| Energy contribution | -14.46 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
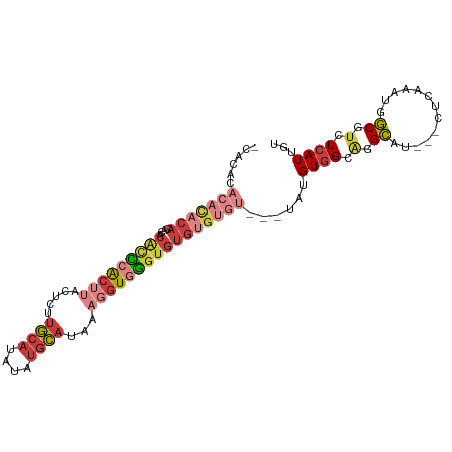
>dm3.chr2L 17619062 101 + 23011544 GUUUUCCCACACACACACACAUAGAGAGACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGAAGGCAU---CUCAAAUGGCGUC (((((((.(((.((((((.(.....)..((((((((((.....((((....))))...))))))))))))))))---..))))))))))..---............. ( -34.90, z-score = -2.62, R) >droSim1.chr2L 17311114 101 + 22036055 GUUUUCCCACACACACACACACACAGAGACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGCAGGCAU---CUCAAAUGGCGUC ((((..(((((...((((((.(.....)((((((((((.....((((....))))...))))))))))))))))---..))))).))))..---............. ( -33.00, z-score = -1.78, R) >droSec1.super_7 1259840 97 + 3727775 GUUUUCCCACACACACACACA----GAGACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGCAGGCAU---CUCAAAUGGCGUC ((((..(((((...((((((.----...((((((((((.....((((....))))...))))))))))))))))---..))))).))))..---............. ( -33.70, z-score = -2.29, R) >droYak2.chr2R 17659766 91 - 21139217 GUUUUCCCACACACACACACA----------CCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGAAGGCAU---CCCAAAUGGCGUC (((((((.(((.(((((((((----------(((((((.....((((....))))...))))))))))))))))---..))))))))))..---.((....)).... ( -42.00, z-score = -5.39, R) >droEre2.scaffold_4845 15957660 97 - 22589142 GUUUUCCCCCACACACACACA----GGCACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGCAGGCAU---CCCAAAUGGCGUC ........(((((.....(((----.((((((((((((.....((((....))))...)))))))))))).)))---..)))))..(.(((---.....))).)... ( -34.80, z-score = -1.86, R) >droAna3.scaffold_12916 12919915 88 - 16180835 ---CCUCCCCACUCAGCCUCG-----------UUACACACACUUGCAUAU--GCAUAAAGGUGAGUGUGCGGGAGUGUAUGUGGCAGGCAU---CUCAAACGGCGAC ---............((((.(-----------(((((.((((((((((((--.(((....))).))))))..)))))).))))))))))..---............. ( -28.90, z-score = -0.96, R) >droPer1.super_8 1947378 96 - 3966273 ------UGUCGCGGUGCCACA----GGAACCCGCGAAAAAGUCACCAAAUCAAGGGAAAGAUAUCACCGAAGGUUGAUG-GUGGCGGGGAUACCCUUCAAUGUCGUC ------..((((((..((...----))...))))))....(((((((((((..((((......)).))...))))..))-)))))(((....)))............ ( -30.20, z-score = -0.98, R) >consensus GUUUUCCCACACACACACACA____GAGACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU___UAUGUGGCAGGCAU___CUCAAAUGGCGUC .......(((((((((................((((((.....(((......)))...))))))))))))))).............(.(((........))).)... (-14.19 = -14.46 + 0.27)

| Location | 17,619,062 – 17,619,163 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 70.65 |
| Shannon entropy | 0.56897 |
| G+C content | 0.49419 |
| Mean single sequence MFE | -35.41 |
| Consensus MFE | -14.99 |
| Energy contribution | -14.43 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17619062 101 - 23011544 GACGCCAUUUGAG---AUGCCUUCCACAUA---ACACACACACCCACCUUUAUGCAUAUAUGCAAGAGUAAGUGGGUGUCUCUCUAUGUGUGUGUGUGUGGGAAAAC .............---.....(((((((((---((((((((((((((((((.((((....))))))))...))))))))........)))))).))))))))).... ( -35.60, z-score = -2.94, R) >droSim1.chr2L 17311114 101 - 22036055 GACGCCAUUUGAG---AUGCCUGCCACAUA---ACACACACACCCACCUUUAUGCAUAUAUGCAAGAGUAAGUGGGUGUCUCUGUGUGUGUGUGUGUGUGGGAAAAC .............---...((..(((((((---((((((((((((((((((.((((....))))))))...))))))))....)))))).)))))).)..))..... ( -38.70, z-score = -2.94, R) >droSec1.super_7 1259840 97 - 3727775 GACGCCAUUUGAG---AUGCCUGCCACAUA---ACACACACACCCACCUUUAUGCAUAUAUGCAAGAGUAAGUGGGUGUCUC----UGUGUGUGUGUGUGGGAAAAC .............---...((..(((((((---.((((.((((((((((((.((((....))))))))...))))))))...----)))))))))).)..))..... ( -35.60, z-score = -2.74, R) >droYak2.chr2R 17659766 91 + 21139217 GACGCCAUUUGGG---AUGCCUUCCACAUA---ACACACACACCCACCUUUAUGCAUAUAUGCAAGAGUAAGUGGG----------UGUGUGUGUGUGUGGGAAAAC ....((....)).---.....(((((((((---.(((((((((((((((((.((((....))))))))...)))))----------))))))))))))))))).... ( -43.00, z-score = -5.31, R) >droEre2.scaffold_4845 15957660 97 + 22589142 GACGCCAUUUGGG---AUGCCUGCCACAUA---ACACACACACCCACCUUUAUGCAUAUAUGCAAGAGUAAGUGGGUGUGCC----UGUGUGUGUGUGGGGGAAAAC ....((....)).---....((.(((((((---((((((((((((((((((.((((....))))))))...)))))))))..----))))).))))))).))..... ( -39.70, z-score = -2.66, R) >droAna3.scaffold_12916 12919915 88 + 16180835 GUCGCCGUUUGAG---AUGCCUGCCACAUACACUCCCGCACACUCACCUUUAUGC--AUAUGCAAGUGUGUGUAA-----------CGAGGCUGAGUGGGGAGG--- ((.((.((.....---..))..)).)).....(((((...(((((((((((((((--((((....))))))))).-----------.)))).))))))))))).--- ( -29.30, z-score = -1.00, R) >droPer1.super_8 1947378 96 + 3966273 GACGACAUUGAAGGGUAUCCCCGCCAC-CAUCAACCUUCGGUGAUAUCUUUCCCUUGAUUUGGUGACUUUUUCGCGGGUUCC----UGUGGCACCGCGACA------ .....((..((((((((((.(((....-..........))).))))))))))...))....(....)....(((((((((..----...))).))))))..------ ( -25.94, z-score = -0.18, R) >consensus GACGCCAUUUGAG___AUGCCUGCCACAUA___ACACACACACCCACCUUUAUGCAUAUAUGCAAGAGUAAGUGGGUGUCUC____UGUGUGUGUGUGUGGGAAAAC ..................................(((((((((.(((((((.(((......)))))))...................))).)))))))))....... (-14.99 = -14.43 + -0.56)


| Location | 17,619,074 – 17,619,170 |
|---|---|
| Length | 96 |
| Sequences | 7 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Shannon entropy | 0.51432 |
| G+C content | 0.46740 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -12.91 |
| Energy contribution | -16.10 |
| Covariance contribution | 3.19 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17619074 96 + 23011544 ACACACACAUAGAGAGACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGAAGGCAU---CUCAAAUGGCGUCUCAUUGU .....((((((((.(.((((((((((.....((((....))))...)))))))))).).).)---))))))((.(((..---(......)..)))))..... ( -31.50, z-score = -1.95, R) >droSim1.chr2L 17311126 96 + 22036055 ACACACACACACAGAGACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGCAGGCAU---CUCAAAUGGCGUCUCAUUGU .((((.((((((....((((((((((.....((((....))))...))))))))))))))))---..))))(.((((..---(......)..)))))..... ( -32.90, z-score = -1.92, R) >droSec1.super_7 1259848 96 + 3727775 ACACACACACACAGAGACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGCAGGCAU---CUCAAAUGGCGUCUCAUUGU .((((.((((((....((((((((((.....((((....))))...))))))))))))))))---..))))(.((((..---(......)..)))))..... ( -32.90, z-score = -1.92, R) >droYak2.chr2R 17659774 90 - 21139217 ------ACACACACACACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGAAGGCAU---CCCAAAUGGCGUCUCAUUGU ------.(((((((((((((((((((.....((((....))))...))))))))))))))))---...)))((.(((..---.((....)).)))))..... ( -36.90, z-score = -3.71, R) >droEre2.scaffold_4845 15957668 96 - 22589142 CCACACACACACAGGCACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU---UAUGUGGCAGGCAU---CCCAAAUGGCGUCUCAUUGU (((((.....(((.((((((((((((.....((((....))))...)))))))))))).)))---..))))).((((..---.((....)).))))...... ( -35.50, z-score = -1.95, R) >droAna3.scaffold_12916 12919922 88 - 16180835 --------ACUCAGCCUCGUU-ACACACACUUGCAUA--UGCAUAAAGGUGAGUGUGCGGGAGUGUAUGUGGCAGGCAU---CUCAAACGGCGACUCAUUGU --------.....((((.(((-(((.(((((((((((--(.(((....))).))))))..)))))).))))))))))..---.................... ( -28.90, z-score = -1.20, R) >droPer1.super_8 1947390 91 - 3966273 ----------ACAGGAACCCGCGAAAAAGUCACCAAAUCAAGGGAAAGAUAUCACCGAAGGUUGAUG-GUGGCGGGGAUACCCUUCAAUGUCGUCUGAUUUU ----------.(((......((((....(((((((((((..((((......)).))...))))..))-)))))(((....))).......)))))))..... ( -23.50, z-score = -0.10, R) >consensus _CACACACACACAGACACACCCACUUACUCUUGCAUAUAUGCAUAAAGGUGGGUGUGUGUGU___UAUGUGGCAGGCAU___CUCAAAUGGCGUCUCAUUGU ......(((((((....(((((((((.....((((....))))...))))))))))))))))....(((.(((.(.(((........))).)))).)))... (-12.91 = -16.10 + 3.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:29 2011