| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,612,817 – 17,612,912 |
| Length | 95 |
| Max. P | 0.561415 |

| Location | 17,612,817 – 17,612,912 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.00 |
| Shannon entropy | 0.43047 |
| G+C content | 0.45175 |
| Mean single sequence MFE | -29.71 |
| Consensus MFE | -11.51 |
| Energy contribution | -12.14 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.561415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17612817 95 + 23011544 -----------------GGUGGGAAUGCC-AAUGCCGCUCAAUAAAUGUCAUUACAUUGUCAAGUGCAUAUUAGCGCCAUAAUUGAACGGCGUGAGCAGAAUUUUCCCCGCCA- -----------------((((((..(((.-.((((((.(((((.(((((....))))).....((((......))))....))))).))))))..)))((....)))))))).- ( -34.40, z-score = -3.50, R) >droSim1.chr2L 17304332 95 + 22036055 -----------------GGUGGGAAUGCC-AAUGCCGCUCAAUAAAUGUCAUUACAUUGUCAAGUGCAUAUUAGCGCCAUAAUUGAACGGCGUGAGCAGAAUUUUCCCCGCCA- -----------------((((((..(((.-.((((((.(((((.(((((....))))).....((((......))))....))))).))))))..)))((....)))))))).- ( -34.40, z-score = -3.50, R) >droSec1.super_7 1253630 95 + 3727775 -----------------GGUGGAAAUGCC-AAUGCCGCUCAAUAAAUGUCAUUACAUUGUCAAGUGCAUAUUAGCGCCAUAAUUGAACGGCGUGAGCAGAAUUUUCCUCGCCA- -----------------(((((((((((.-.((((((.(((((.(((((....))))).....((((......))))....))))).))))))..)))....)))))..))).- ( -30.50, z-score = -2.69, R) >droYak2.chr2R 17653370 95 - 21139217 -----------------GGAGGGAAUGCC-AAUGCCGCUCAAUAAAUGUCAUUACAUUGUCAAGUGCAUAUUAGCGCCAUAAUUGAACGGCGUGCACAGAAUUUUCCCCGCCA- -----------------((.((((((((.-.((((((.(((((.(((((....))))).....((((......))))....))))).))))))))).......)))))..)).- ( -28.71, z-score = -1.92, R) >droEre2.scaffold_4845 15951105 95 - 22589142 -----------------GGCGGGAAUGCC-AAUGCCGCUCAAUAAAUGUCAUUACAUUGUCAAGUGCAUAUUAGCGCCAUAAUUGAACGGCGUGAACAGAAUUUUCCCCGCCA- -----------------((((((..((..-.((((((.(((((.(((((....))))).....((((......))))....))))).))))))...))((....)))))))).- ( -32.20, z-score = -2.95, R) >droAna3.scaffold_12916 3265128 97 + 16180835 ----------------GGGAGGAAAUGCC-AAUGCCGCUCAAUAAAUGUCAUUACAUUGUCAAGUGCAUAUUAGCGCCAUAAUUGAGCGGCGUGAGCGGAAUUUUCCCAGCCAG ----------------(((((((..(((.-.((((((((((((.(((((....))))).....((((......))))....))))))))))))..)))...)))))))...... ( -39.80, z-score = -4.96, R) >droWil1.scaffold_180764 869772 94 + 3949147 ----------AACCACGGCCAAUGGGGAAAAAUGGAGCUCAAUAAAUGUCAUUACAUUGUCAAGUGCAUAUUAGCGCCAUAAUUGCGCCGCGUGAGUUGAAUUU---------- ----------(((((((((((((..((...((((..(((.....(((((....)))))....)))...))))....))...)))).)))..))).)))......---------- ( -18.40, z-score = 1.30, R) >droVir3.scaffold_12963 3650190 109 - 20206255 GAAGCUGAAGAAGCAGCAGCAGCAGCAGAAGAUGCCGCUCAAUAAAUGUCAUUACAUUGUCAAGUGCAUAUUAGAGCCAUAAUUGCGCGACGUGCGCAGCAUUUUCAGA----- ...((((......)))).((....)).((((((((.((.((...(((((....)))))(((..(((((.((((......)))))))))))).)).)).))))))))...----- ( -31.50, z-score = -1.15, R) >droGri2.scaffold_15252 13975683 80 + 17193109 -----------------AGUCGUCGCACA-GAUUUCGCUCAAUAAAUGUCAUUAGAUUGUCAAGUGCAUAUUAGAGCCAUAAUUGCGAGACGUGCUCA---------------- -----------------.(((.(((((..-.(((..((((((((..((.((((.(.....).)))))))))).))))...))))))))))).......---------------- ( -17.50, z-score = -0.38, R) >consensus _________________GGUGGGAAUGCC_AAUGCCGCUCAAUAAAUGUCAUUACAUUGUCAAGUGCAUAUUAGCGCCAUAAUUGAACGGCGUGAGCAGAAUUUUCCCCGCCA_ ...............................((((((..((((...((.((......)).)).((((......))))....))))..))))))..................... (-11.51 = -12.14 + 0.63)

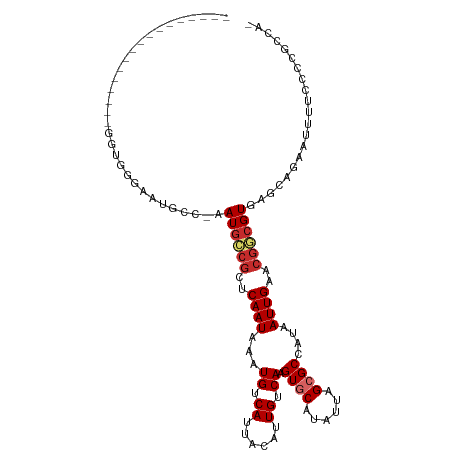
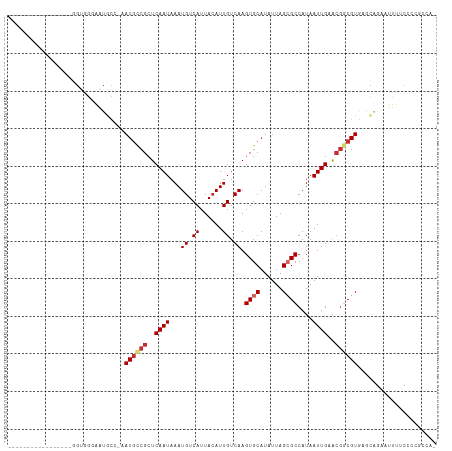
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:27 2011