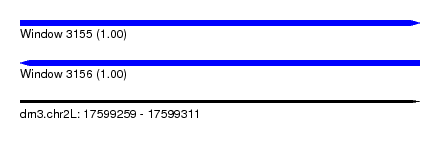
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,599,259 – 17,599,311 |
| Length | 52 |
| Max. P | 0.999876 |

| Location | 17,599,259 – 17,599,311 |
|---|---|
| Length | 52 |
| Sequences | 6 |
| Columns | 53 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Shannon entropy | 0.40653 |
| G+C content | 0.52929 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -11.99 |
| Energy contribution | -12.77 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.39 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
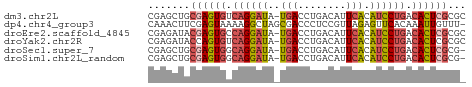
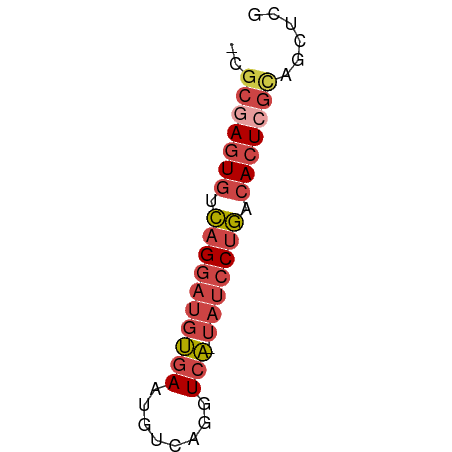
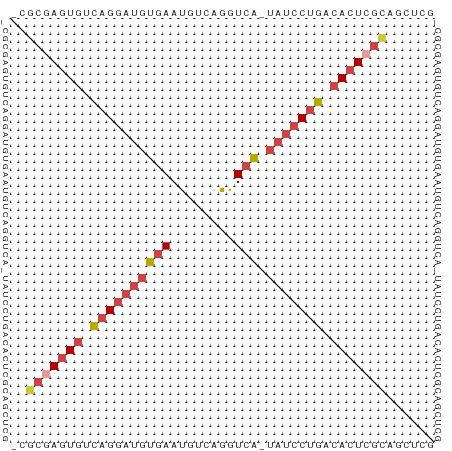
>dm3.chr2L 17599259 52 + 23011544 CGAGCUGCGAGUGUCAGGAUA-UGACCUGACAUUCACAUCCUGACACUCGCGC ...((.((((((((((((((.-(((........))).)))))))))))))))) ( -29.80, z-score = -6.91, R) >dp4.chr4_group3 859361 52 - 11692001 CAAACUUCGAGUAAAAGGCUAGCGACCCUCCGUUAGAGUUCACAAAUUGUUU- .((((.....((.((...((((((......))))))..)).)).....))))- ( -7.60, z-score = -0.24, R) >droEre2.scaffold_4845 15937284 52 - 22589142 CGAGAUACGAGUGCCAGGAUA-UGACCUGACAUUCACAUCCUGACACUCGCGC .......((((((.((((((.-(((........))).)))))).))))))... ( -20.10, z-score = -4.71, R) >droYak2.chr2R 17639537 52 - 21139217 CGAGAUACCAGUGUCAGGAUA-UGACCUGACAUUCACAUCCUGACACUCGCGC .........(((((((((((.-(((........))).)))))))))))..... ( -19.30, z-score = -3.95, R) >droSec1.super_7 1240562 51 + 3727775 CGAGCUGCGAGUGGCAGGAUA-UGACCUGACAUUCACAUCCUGACACUCGCG- ......(((((((.((((((.-(((........))).)))))).))))))).- ( -24.30, z-score = -5.25, R) >droSim1.chr2L_random 656304 51 + 909653 CGAGCUGCGAGUGGCAGGAUA-UGACCUGACAUUCACAUCCUGACACUCGCG- ......(((((((.((((((.-(((........))).)))))).))))))).- ( -24.30, z-score = -5.25, R) >consensus CGAGCUGCGAGUGGCAGGAUA_UGACCUGACAUUCACAUCCUGACACUCGCG_ .......((((((.(((((...(((........)))..))))).))))))... (-11.99 = -12.77 + 0.78)
| Location | 17,599,259 – 17,599,311 |
|---|---|
| Length | 52 |
| Sequences | 6 |
| Columns | 53 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Shannon entropy | 0.40653 |
| G+C content | 0.52929 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -16.07 |
| Energy contribution | -17.35 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -5.26 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
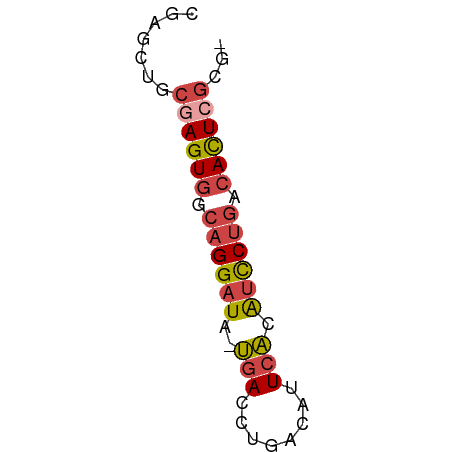
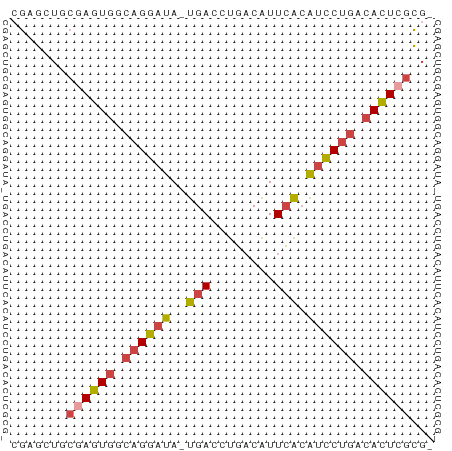
>dm3.chr2L 17599259 52 - 23011544 GCGCGAGUGUCAGGAUGUGAAUGUCAGGUCA-UAUCCUGACACUCGCAGCUCG ((((((((((((((((((((........)))-))))))))))))))).))... ( -35.00, z-score = -7.71, R) >dp4.chr4_group3 859361 52 + 11692001 -AAACAAUUUGUGAACUCUAACGGAGGGUCGCUAGCCUUUUACUCGAAGUUUG -((((.....((((((((.....)))(((.....))).))))).....)))). ( -7.50, z-score = 0.56, R) >droEre2.scaffold_4845 15937284 52 + 22589142 GCGCGAGUGUCAGGAUGUGAAUGUCAGGUCA-UAUCCUGGCACUCGUAUCUCG ..((((((((((((((((((........)))-)))))))))))))))...... ( -30.80, z-score = -6.92, R) >droYak2.chr2R 17639537 52 + 21139217 GCGCGAGUGUCAGGAUGUGAAUGUCAGGUCA-UAUCCUGACACUGGUAUCUCG ..((.(((((((((((((((........)))-)))))))))))).))...... ( -27.60, z-score = -5.40, R) >droSec1.super_7 1240562 51 - 3727775 -CGCGAGUGUCAGGAUGUGAAUGUCAGGUCA-UAUCCUGCCACUCGCAGCUCG -.(((((((.((((((((((........)))-))))))).)))))))...... ( -28.80, z-score = -6.04, R) >droSim1.chr2L_random 656304 51 - 909653 -CGCGAGUGUCAGGAUGUGAAUGUCAGGUCA-UAUCCUGCCACUCGCAGCUCG -.(((((((.((((((((((........)))-))))))).)))))))...... ( -28.80, z-score = -6.04, R) >consensus _CGCGAGUGUCAGGAUGUGAAUGUCAGGUCA_UAUCCUGACACUCGCAGCUCG ..(((((((.((((((((((........))).))))))).)))))))...... (-16.07 = -17.35 + 1.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:26 2011