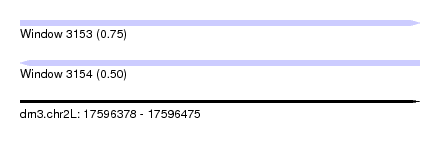
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,596,378 – 17,596,475 |
| Length | 97 |
| Max. P | 0.745215 |

| Location | 17,596,378 – 17,596,475 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 88.01 |
| Shannon entropy | 0.20440 |
| G+C content | 0.39852 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -13.84 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
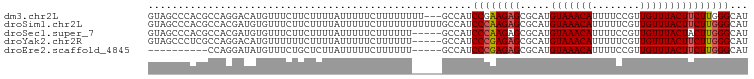
>dm3.chr2L 17596378 97 + 23011544 GUAGCCCACGCCAGGACAUGUUUCUUCUUUUAUUUUUCUUUUUUUU---GCCAUCCGAAGAGCGCAUGUAAACAUUUUCCGUUGUUUACUUCUUGGGCAU ...(((((....((((.((((((((((...................---.......)))))).))))(((((((........)))))))))))))))).. ( -21.27, z-score = -2.23, R) >droSim1.chr2L 17300011 100 + 22036055 GUAGCCCACGCCACGAUGUGUUUCUUCUUUUAUUUUUCUUUUUUUUUUUGCCAUCCCAAGAGCGCAUGUAAACAUUUUUCGUUGUUUACUUCUUGGGCAU ...(((((.......(((((((.(((...............................))))))))))(((((((........)))))))....))))).. ( -20.68, z-score = -2.87, R) >droSec1.super_7 1237697 95 + 3727775 GUAGCCCACGCCACGAUGUGUUUCUUCUUUUAUUUUUCUUUUUU-----GCCAUCCCAAGAGCGCAUGUAAACAUUUUCCGUUGUUUACUACUUGGGCAU ...(((((.......(((((((.(((..................-----........))))))))))(((((((........)))))))....))))).. ( -20.87, z-score = -2.65, R) >droYak2.chr2R 17636759 95 - 21139217 GUAGCCCUCGCCAGGACAUGUUUUUUCUUUUAUUUUUCUUUUUU-----GCCAUCCCGAGAGCGCAUGUAAACAUUUUUCGUUGUUUACUUCUUGGGCAU ...((((........((((((.(((((.................-----........))))).))))))(((((........))))).......)))).. ( -18.21, z-score = -1.29, R) >droEre2.scaffold_4845 15934630 85 - 22589142 ----------CCAGGAUAUGUUUCUGCUCUUAUUUUUCUUUUUU-----GCCAUCCCGAGAGCGCAUGUAAACAUUUUCCGUUGUUUACUUCUUGGGCAU ----------((((((.((((....((((((.............-----........))))))))))(((((((........))))))).)))))).... ( -20.10, z-score = -2.43, R) >consensus GUAGCCCACGCCAGGAUAUGUUUCUUCUUUUAUUUUUCUUUUUU_____GCCAUCCCAAGAGCGCAUGUAAACAUUUUCCGUUGUUUACUUCUUGGGCAU ......................................................((((((((.....(((((((........)))))))))))))))... (-13.84 = -14.00 + 0.16)


| Location | 17,596,378 – 17,596,475 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.01 |
| Shannon entropy | 0.20440 |
| G+C content | 0.39852 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -15.19 |
| Energy contribution | -16.91 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr2L 17596378 97 - 23011544 AUGCCCAAGAAGUAAACAACGGAAAAUGUUUACAUGCGCUCUUCGGAUGGC---AAAAAAAAGAAAAAUAAAAGAAGAAACAUGUCCUGGCGUGGGCUAC ..((((.....(((((((........))))))).(((..((....))..))---)..........................((((....))))))))... ( -21.60, z-score = -1.63, R) >droSim1.chr2L 17300011 100 - 22036055 AUGCCCAAGAAGUAAACAACGAAAAAUGUUUACAUGCGCUCUUGGGAUGGCAAAAAAAAAAAGAAAAAUAAAAGAAGAAACACAUCGUGGCGUGGGCUAC ..((((.....(((((((........)))))))..(((((...(.((((.................................)))).))))))))))... ( -22.31, z-score = -2.52, R) >droSec1.super_7 1237697 95 - 3727775 AUGCCCAAGUAGUAAACAACGGAAAAUGUUUACAUGCGCUCUUGGGAUGGC-----AAAAAAGAAAAAUAAAAGAAGAAACACAUCGUGGCGUGGGCUAC ..((((.....(((((((........)))))))..(((((...(.((((..-----..........................)))).))))))))))... ( -22.49, z-score = -1.90, R) >droYak2.chr2R 17636759 95 + 21139217 AUGCCCAAGAAGUAAACAACGAAAAAUGUUUACAUGCGCUCUCGGGAUGGC-----AAAAAAGAAAAAUAAAAGAAAAAACAUGUCCUGGCGAGGGCUAC ..((((.....(((((((........)))))))...(((...((((((...-----...........................))))))))).))))... ( -22.21, z-score = -2.36, R) >droEre2.scaffold_4845 15934630 85 + 22589142 AUGCCCAAGAAGUAAACAACGGAAAAUGUUUACAUGCGCUCUCGGGAUGGC-----AAAAAAGAAAAAUAAGAGCAGAAACAUAUCCUGG---------- ....(((.((.(((((((........)))))))....(((((........(-----......).......))))).........)).)))---------- ( -14.26, z-score = -0.33, R) >consensus AUGCCCAAGAAGUAAACAACGGAAAAUGUUUACAUGCGCUCUUGGGAUGGC_____AAAAAAGAAAAAUAAAAGAAGAAACAUAUCCUGGCGUGGGCUAC ..((((.....(((((((........)))))))..(((((...((((((.................................)))))))))))))))... (-15.19 = -16.91 + 1.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:25 2011