| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,592,558 – 17,592,657 |
| Length | 99 |
| Max. P | 0.857998 |

| Location | 17,592,558 – 17,592,657 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 74.25 |
| Shannon entropy | 0.49771 |
| G+C content | 0.36554 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -9.69 |
| Energy contribution | -11.63 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
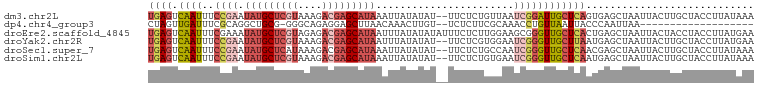
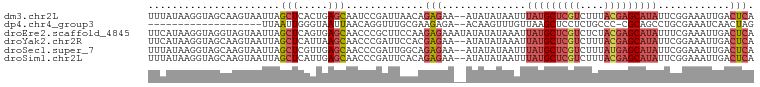
>dm3.chr2L 17592558 99 + 23011544 UGAGUCAAUUUCCGAAUAUGCUCGUAAAGACGAGCAUAAAUUAUAUAU--UUCUCUGUUAAUCGGAUUGCUCAGUGAGCUAAUUACUUGCUACCUUAUAAA .(((.(((..(((((.(((((((((....)))))))))(((.....))--)..........)))))))))))((..((.......))..)).......... ( -25.00, z-score = -3.21, R) >dp4.chr4_group3 851393 79 - 11692001 CUAGUUGAUUUCGCAGGCUGCG-GGGCAGAGGAGCUUAACAAACUUGU--UCUCUUCGCAAACCUGUUAAUUACCCAAUUAA------------------- .((((((..((.(((((.((((-(((....(((((...........))--))))))))))..))))).)).....)))))).------------------- ( -20.00, z-score = -0.97, R) >droEre2.scaffold_4845 15930860 101 - 22589142 UGAGUCAAUUUCGAAAUAUGCUCGUAGAGACGAGCAUAAUUUAUAUAUAUUUCUCUUGGAAGCGGGUUGCUCACUGAGCUAAUUACUACCUACCUUAUGAA .(((.........((((((((((((....)))))))).))))..........)))..((.((..(...((((...))))...)..)).))........... ( -21.91, z-score = -1.04, R) >droYak2.chr2R 17632917 99 - 21139217 UGAGUCAAUUUCCGAAUAUGCUCGUAAAGACGAGCAUAAUUUAUAUAU--UUCUCGUGGAAUCGGGUUGCUUAAUGAGCUAAUUACUUGCUACCUUAUGAA ((((.((((..((((.(((((((((....))))))))).........(--(((....))))))))))))))))...(((.........))).......... ( -24.30, z-score = -2.14, R) >droSec1.super_7 1233814 99 + 3727775 UGAGUCAAUUUCCGAAUAUGCUCAUAAAGACGAGCAUAAAUUAUAUAU--UUCUCUGCCAAUCGGGUUGCUCAACGAGCUAAUUACUUGCUACCUUAUAAA ((((.((((..((((.(((((((........)))))))(((.....))--)..........))))))))))))...(((.........))).......... ( -19.40, z-score = -1.58, R) >droSim1.chr2L 17296185 99 + 22036055 UGAGUCAAUUUCCGAAUAUGCUCGUAAAGACGAGCAUAAAUUAUAUAU--UUCUCUGUGAAUCGGGUUGCUCAAUGAGCUAAUUACUUGCUACCUUAUAAA ((((.((((..((((.(((((((((....)))))))))..(((((...--.....))))).))))))))))))...(((.........))).......... ( -26.00, z-score = -3.41, R) >consensus UGAGUCAAUUUCCGAAUAUGCUCGUAAAGACGAGCAUAAAUUAUAUAU__UUCUCUGGGAAUCGGGUUGCUCAAUGAGCUAAUUACUUGCUACCUUAUAAA ((((.((((..((((.(((((((((....))))))))).......................))))))))))))............................ ( -9.69 = -11.63 + 1.95)
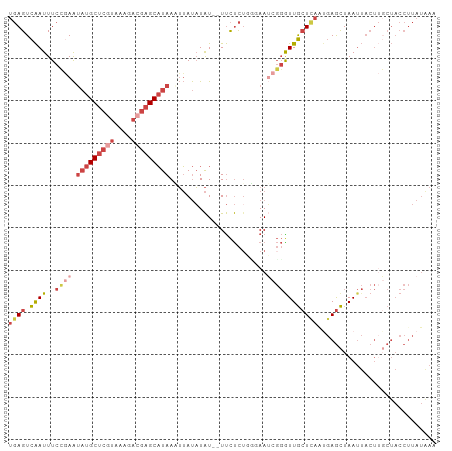
| Location | 17,592,558 – 17,592,657 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 74.25 |
| Shannon entropy | 0.49771 |
| G+C content | 0.36554 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -10.03 |
| Energy contribution | -11.17 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
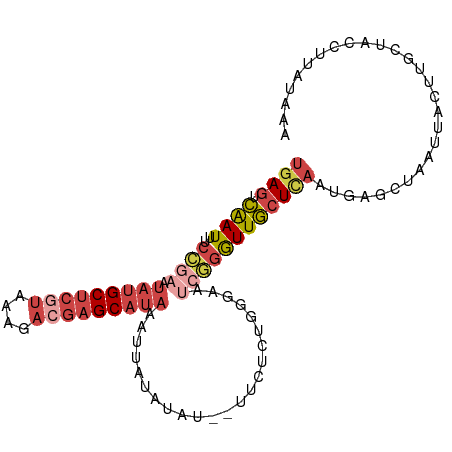
>dm3.chr2L 17592558 99 - 23011544 UUUAUAAGGUAGCAAGUAAUUAGCUCACUGAGCAAUCCGAUUAACAGAGAA--AUAUAUAAUUUAUGCUCGUCUUUACGAGCAUAUUCGGAAAUUGACUCA .......((((((((....)).))).)))(((((((((((.......(...--.)........(((((((((....))))))))).)))))..))).))). ( -25.00, z-score = -3.13, R) >dp4.chr4_group3 851393 79 + 11692001 -------------------UUAAUUGGGUAAUUAACAGGUUUGCGAAGAGA--ACAAGUUUGUUAAGCUCCUCUGCCC-CGCAGCCUGCGAAAUCAACUAG -------------------....((((....((..((((((.(((.((((.--...((((.....)))).))))....-)))))))))..)).)))).... ( -15.60, z-score = 0.21, R) >droEre2.scaffold_4845 15930860 101 + 22589142 UUCAUAAGGUAGGUAGUAAUUAGCUCAGUGAGCAACCCGCUUCCAAGAGAAAUAUAUAUAAAUUAUGCUCGUCUCUACGAGCAUAUUUCGAAAUUGACUCA ...........((.(((.....((((...)))).....))).))..(((..............(((((((((....)))))))))..(((....)))))). ( -21.50, z-score = -1.40, R) >droYak2.chr2R 17632917 99 + 21139217 UUCAUAAGGUAGCAAGUAAUUAGCUCAUUAAGCAACCCGAUUCCACGAGAA--AUAUAUAAAUUAUGCUCGUCUUUACGAGCAUAUUCGGAAAUUGACUCA .......(((.((...((((......)))).)).)))(((((((...(...--.)........(((((((((....)))))))))...)).)))))..... ( -19.70, z-score = -1.81, R) >droSec1.super_7 1233814 99 - 3727775 UUUAUAAGGUAGCAAGUAAUUAGCUCGUUGAGCAACCCGAUUGGCAGAGAA--AUAUAUAAUUUAUGCUCGUCUUUAUGAGCAUAUUCGGAAAUUGACUCA .....((.(.(((((....)).)))).))((((((.((((.......(...--.)........(((((((((....))))))))).))))...))).))). ( -19.80, z-score = -0.38, R) >droSim1.chr2L 17296185 99 - 22036055 UUUAUAAGGUAGCAAGUAAUUAGCUCAUUGAGCAACCCGAUUCACAGAGAA--AUAUAUAAUUUAUGCUCGUCUUUACGAGCAUAUUCGGAAAUUGACUCA ........(.(((((....)).))))..(((((((.(((((((.....)))--..........(((((((((....))))))))).))))...))).)))) ( -22.80, z-score = -2.53, R) >consensus UUUAUAAGGUAGCAAGUAAUUAGCUCAUUGAGCAACCCGAUUCCCAGAGAA__AUAUAUAAUUUAUGCUCGUCUUUACGAGCAUAUUCGGAAAUUGACUCA ......................(((.....))).............(((..............(((((((((....)))))))))............))). (-10.03 = -11.17 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:23 2011