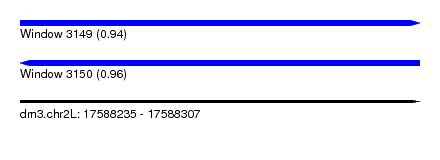
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,588,235 – 17,588,307 |
| Length | 72 |
| Max. P | 0.963834 |

| Location | 17,588,235 – 17,588,307 |
|---|---|
| Length | 72 |
| Sequences | 12 |
| Columns | 88 |
| Reading direction | forward |
| Mean pairwise identity | 81.69 |
| Shannon entropy | 0.37789 |
| G+C content | 0.39307 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.23 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17588235 72 + 23011544 ---------GAAAAUAUUUGGCAUGUGCCAUUUA-UGGCAUUGUUCCAGC-UUUGAACAAU-GCGAUACCAUAAUUUCCAGGCA---- ---------(((((((..((((....))))..))-).(((((((((....-...)))))))-))..........))))......---- ( -20.00, z-score = -1.81, R) >droSim1.chr2L 17291952 72 + 22036055 ---------GAAAAUAUUUGGCAUGUGCCAUUUA-UGGCAUUGUUCCAGC-UUUGAACAAU-GCGAUACCAUAAUUUCCAGGCA---- ---------(((((((..((((....))))..))-).(((((((((....-...)))))))-))..........))))......---- ( -20.00, z-score = -1.81, R) >droSec1.super_7 1229551 72 + 3727775 ---------GAAAAUAUUUGGCAUGUGCCAUUUA-UGGCAUUGUUCCAGC-UUUGAACAAU-GCGAUACCAUAAUUUCCAGGCA---- ---------(((((((..((((....))))..))-).(((((((((....-...)))))))-))..........))))......---- ( -20.00, z-score = -1.81, R) >droYak2.chr2R 17628663 72 - 21139217 ---------GAAAAUAUUUGGCAUGUGCCAUUUA-UGGCAUUGUUCCAGC-UUUGAACAAU-GCGAUACCAUAAUUUCCAGGCA---- ---------(((((((..((((....))))..))-).(((((((((....-...)))))))-))..........))))......---- ( -20.00, z-score = -1.81, R) >droEre2.scaffold_4845 15926676 72 - 22589142 ---------GAAAAUAUUUGGCAUGUGCCAUUUA-UGGCAUUGUUCCAGC-UUUGAACAAU-GCGAUACCAUAAUUUUCAGGCA---- ---------((((((...((((....))))..((-(.(((((((((....-...)))))))-)).))).....)))))).....---- ( -22.70, z-score = -2.94, R) >droAna3.scaffold_12916 12891571 72 - 16180835 ---------GAAAAUAUUUGGCAUGUGCCAUUUA-UGGCAUUGUUCCAAG-UUGGAACAAU-GCGAUACCAUAGUUUCCAGGCA---- ---------((((.(((.((((....))))..((-(.((((((((((...-..))))))))-)).)))..))).))))......---- ( -24.50, z-score = -3.15, R) >dp4.chr4_group3 847368 73 - 11692001 --------GAAAUAUAUUUGGCAUGUGCCAUUUA-UGGCAUUGUUCCAAC-UUUGAACAAU-GCGAUACCAUAAUUUCCAGGCA---- --------(((((.....((((....))))..((-(.(((((((((....-...)))))))-)).))).....)))))......---- ( -20.90, z-score = -2.59, R) >droPer1.super_8 1910752 73 - 3966273 --------GAAAUAUAUUUGGCAUGUGCCAUUUA-UGGCAUUGUUCCAAC-UUUGAACAAU-GCGAUACCAUAAUUUCCAGGCA---- --------(((((.....((((....))))..((-(.(((((((((....-...)))))))-)).))).....)))))......---- ( -20.90, z-score = -2.59, R) >droWil1.scaffold_180764 3040729 74 - 3949147 ---------GAAAAUAUUUGGCAUGUGCCAUUUA-UGGCAUUGUUCCAACAUCUGAACAAUUGCGAUACCAUAAUUUUCAGGCA---- ---------((((((...((((....))))..((-(.(((((((((........)))))).))).))).....)))))).....---- ( -19.20, z-score = -1.95, R) >droVir3.scaffold_12963 16521758 61 + 20206255 ---------AAAAAUGUUUGGCAUGUGCCAUUCGCUGGCAUUGUGCCCAC-UUUAAACAAU-GCGAUGC-ACA--------------- ---------......(..((((....))))..)(((.(((((((......-.....)))))-)).).))-...--------------- ( -18.00, z-score = -1.09, R) >droMoj3.scaffold_6500 21449344 77 + 32352404 ---------GAAAAUGUUUGGCAUGUGCCAAACGCUGGCAUUGUUUUCAC-UUUGAACAAU-GCGAUACCAUAAUUUGCAGCACAACA ---------......(((((((....)))))))(((((((((((((....-...)))))))-)).....((.....))))))...... ( -26.10, z-score = -3.44, R) >droGri2.scaffold_15252 13932840 83 + 17193109 UACAACACUGGCCCCGUUGAAACAACGCCAUUGCCUGGCAUUGUUCCCAACUUUGAACAAU-GCGAUAACAUAAUUUACAGACA---- ..((.((.((((...((((...)))))))).))..))(((((((((........)))))))-))....................---- ( -18.90, z-score = -2.84, R) >consensus _________GAAAAUAUUUGGCAUGUGCCAUUUA_UGGCAUUGUUCCAAC_UUUGAACAAU_GCGAUACCAUAAUUUCCAGGCA____ ..............((..((((....))))..))...(((((((((........))))))).))........................ (-12.00 = -12.23 + 0.22)

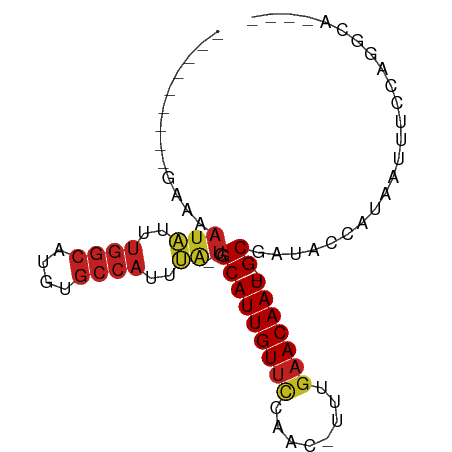
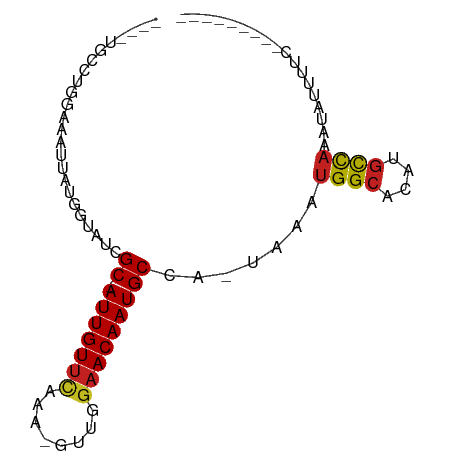
| Location | 17,588,235 – 17,588,307 |
|---|---|
| Length | 72 |
| Sequences | 12 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Shannon entropy | 0.37789 |
| G+C content | 0.39307 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -14.35 |
| Energy contribution | -14.81 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.963834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17588235 72 - 23011544 ----UGCCUGGAAAUUAUGGUAUCGC-AUUGUUCAAA-GCUGGAACAAUGCCA-UAAAUGGCACAUGCCAAAUAUUUUC--------- ----((((..........))))..((-(((((((...-....)))))))))..-....((((....)))).........--------- ( -19.70, z-score = -1.81, R) >droSim1.chr2L 17291952 72 - 22036055 ----UGCCUGGAAAUUAUGGUAUCGC-AUUGUUCAAA-GCUGGAACAAUGCCA-UAAAUGGCACAUGCCAAAUAUUUUC--------- ----((((..........))))..((-(((((((...-....)))))))))..-....((((....)))).........--------- ( -19.70, z-score = -1.81, R) >droSec1.super_7 1229551 72 - 3727775 ----UGCCUGGAAAUUAUGGUAUCGC-AUUGUUCAAA-GCUGGAACAAUGCCA-UAAAUGGCACAUGCCAAAUAUUUUC--------- ----((((..........))))..((-(((((((...-....)))))))))..-....((((....)))).........--------- ( -19.70, z-score = -1.81, R) >droYak2.chr2R 17628663 72 + 21139217 ----UGCCUGGAAAUUAUGGUAUCGC-AUUGUUCAAA-GCUGGAACAAUGCCA-UAAAUGGCACAUGCCAAAUAUUUUC--------- ----((((..........))))..((-(((((((...-....)))))))))..-....((((....)))).........--------- ( -19.70, z-score = -1.81, R) >droEre2.scaffold_4845 15926676 72 + 22589142 ----UGCCUGAAAAUUAUGGUAUCGC-AUUGUUCAAA-GCUGGAACAAUGCCA-UAAAUGGCACAUGCCAAAUAUUUUC--------- ----((((..........))))..((-(((((((...-....)))))))))..-....((((....)))).........--------- ( -19.70, z-score = -2.26, R) >droAna3.scaffold_12916 12891571 72 + 16180835 ----UGCCUGGAAACUAUGGUAUCGC-AUUGUUCCAA-CUUGGAACAAUGCCA-UAAAUGGCACAUGCCAAAUAUUUUC--------- ----((((.(....)...))))..((-((((((((..-...))))))))))..-....((((....)))).........--------- ( -26.70, z-score = -4.92, R) >dp4.chr4_group3 847368 73 + 11692001 ----UGCCUGGAAAUUAUGGUAUCGC-AUUGUUCAAA-GUUGGAACAAUGCCA-UAAAUGGCACAUGCCAAAUAUAUUUC-------- ----((((..........))))..((-(((((((...-....)))))))))..-....((((....))))..........-------- ( -19.70, z-score = -1.90, R) >droPer1.super_8 1910752 73 + 3966273 ----UGCCUGGAAAUUAUGGUAUCGC-AUUGUUCAAA-GUUGGAACAAUGCCA-UAAAUGGCACAUGCCAAAUAUAUUUC-------- ----((((..........))))..((-(((((((...-....)))))))))..-....((((....))))..........-------- ( -19.70, z-score = -1.90, R) >droWil1.scaffold_180764 3040729 74 + 3949147 ----UGCCUGAAAAUUAUGGUAUCGCAAUUGUUCAGAUGUUGGAACAAUGCCA-UAAAUGGCACAUGCCAAAUAUUUUC--------- ----.....((((((..((((((....(((((((........)))))))((((-....))))..))))))...))))))--------- ( -19.60, z-score = -1.89, R) >droVir3.scaffold_12963 16521758 61 - 20206255 ---------------UGU-GCAUCGC-AUUGUUUAAA-GUGGGCACAAUGCCAGCGAAUGGCACAUGCCAAACAUUUUU--------- ---------------(((-((.((((-(((......)-)).(((.....))).))))...)))))..............--------- ( -16.20, z-score = -0.27, R) >droMoj3.scaffold_6500 21449344 77 - 32352404 UGUUGUGCUGCAAAUUAUGGUAUCGC-AUUGUUCAAA-GUGAAAACAAUGCCAGCGUUUGGCACAUGCCAAACAUUUUC--------- ......((((((.....)).....((-(((((((...-..))..)))))))))))(((((((....)))))))......--------- ( -22.40, z-score = -1.61, R) >droGri2.scaffold_15252 13932840 83 - 17193109 ----UGUCUGUAAAUUAUGUUAUCGC-AUUGUUCAAAGUUGGGAACAAUGCCAGGCAAUGGCGUUGUUUCAACGGGGCCAGUGUUGUA ----................(((.((-((((..(...(((((.((((((((((.....)))))))))))))))..)..)))))).))) ( -28.20, z-score = -3.11, R) >consensus ____UGCCUGGAAAUUAUGGUAUCGC_AUUGUUCAAA_GUUGGAACAAUGCCA_UAAAUGGCACAUGCCAAAUAUUUUC_________ .................((((((....(((((((........)))))))((((.....))))..)))))).................. (-14.35 = -14.81 + 0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:22 2011