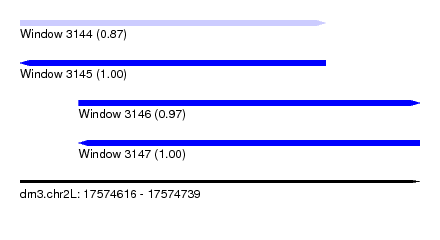
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,574,616 – 17,574,739 |
| Length | 123 |
| Max. P | 0.997041 |

| Location | 17,574,616 – 17,574,710 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.96 |
| Shannon entropy | 0.39973 |
| G+C content | 0.36534 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -18.45 |
| Energy contribution | -18.30 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
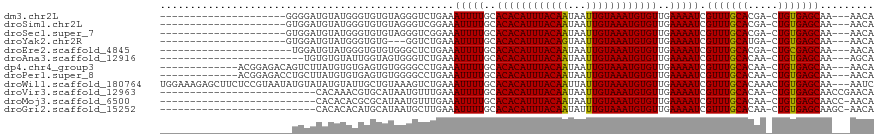
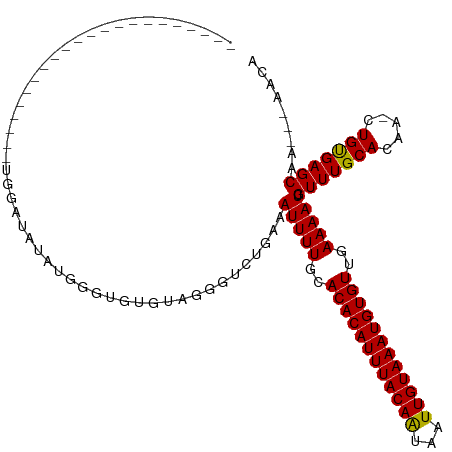
>dm3.chr2L 17574616 94 + 23011544 ---------------------GGGGAUGUAUGGGUGUGUAGGGUCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACGA-CUGUGAGCAA---AACA ---------------------...(...(((((.((((((((...(((..(((((.(((((((((((.....)))))))))))))))).))))))))))).-)))))..)..---.... ( -22.10, z-score = -0.84, R) >droSim1.chr2L 17277837 94 + 22036055 ---------------------GUGGAUGUAUGGGUGUGUAGGGUCGGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACGA-CUGUGAGCAA---AACA ---------------------((.(...(((((.(((((((...(((...(((((.(((((((((((.....)))))))))))))))).))).))))))).-)))))..)..---.)). ( -24.00, z-score = -1.29, R) >droSec1.super_7 1215454 94 + 3727775 ---------------------GUGGAUGUAUGGGUGUGUAGGGUCGGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACGA-CUGUGAGCAA---AACA ---------------------((.(...(((((.(((((((...(((...(((((.(((((((((((.....)))))))))))))))).))).))))))).-)))))..)..---.)). ( -24.00, z-score = -1.29, R) >droYak2.chr2R 17614005 91 - 21139217 ---------------------GUGGAUGUAUGGGUGUG---GGUCUGAAAUUUUGCACACAUUUACAGUAAUUGUAAAUGUGUUGAAAAUCGUUUGCAUGA-CUGUGAGCAA---AACA ---------------------((...((((((.(((..---((........))..))).)))(((((((...((((((((...(....).))))))))..)-))))))))).---.)). ( -23.60, z-score = -1.33, R) >droEre2.scaffold_4845 15912966 93 - 22589142 ----------------------UGGAUGUAUGGGUGUGUGGGCUCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACGA-CUGCGAGCAA---AACA ----------------------..(((....((((......)))).....(((((.(((((((((((.....)))))))))))))))))))(((((((...-.)))))))..---.... ( -23.90, z-score = -1.14, R) >droAna3.scaffold_12916 12878296 91 - 16180835 ------------------------UGUGUGUAUUGGUAGUGGGUCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAA---AGCA ------------------------..(((....((((.(((.(..(((..(((((.(((((((((((.....)))))))))))))))).)))..).))).)-)))...))).---.... ( -21.60, z-score = -0.39, R) >dp4.chr4_group3 833033 102 - 11692001 -------------ACGGAGACAGUCUUAUGUGUGAGUGUGGGGCCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAA---AACA -------------..(....).((((((..(....)..))))))......(((((((((((((((((.....))))))))))).............(((..-..))).))))---)).. ( -27.00, z-score = -1.08, R) >droPer1.super_8 1896219 102 - 3966273 -------------ACGGAGACCUGCUUAUGUGUGAGUGUGGGGCCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAA---AACA -------------..(....).((((((((.((..(((..((...(((..(((((.(((((((((((.....)))))))))))))))).)))))..))).)-))))))))).---.... ( -27.50, z-score = -1.31, R) >droWil1.scaffold_180764 3020211 116 - 3949147 UGGAAAGAGCUUCUCCGUAAUAUGUAUAUGUAUUGCUGUAAAGUCUGAAAUUUUGCACACAUUUACAAUUAUUGUAAAUGUGUUGAAAAUCGUUUGCACAAACUGUGAGCAA---AAUC .....(((.(((..(.(((((((......))))))).)..))))))...((((((((((((((((((.....))))))))))).............(((.....))).))))---))). ( -28.80, z-score = -2.68, R) >droVir3.scaffold_12963 16502557 92 + 20206255 --------------------------CACAAACGUGCAUAAUGUUUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAACCGAACA --------------------------..(((((((.....)))))))...(((((.(((((((((((.....)))))))))))))))).(((.((((.((.-...)).)))).)))... ( -26.10, z-score = -2.65, R) >droMoj3.scaffold_6500 21428995 91 + 32352404 --------------------------CACACACGCGCAUAAUGUUUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAACC-AACA --------------------------.........(((.(((.......))).)))(((((((((((.....)))))))))))........(((..((...-.))..)))....-.... ( -20.90, z-score = -0.83, R) >droGri2.scaffold_15252 13917111 91 + 17193109 --------------------------CACACACAUGCAUAAUGCUUGAAAUUUUGCACACAUUUACAAUAUUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAAGC-AACA --------------------------...............(((((...(((((..(((((((((((.....)))))))))))..))))).(((..((...-.))..)))))))-)... ( -23.50, z-score = -1.47, R) >consensus ______________________UGGAUAUAUGGGUGUGUAGGGUCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA_CUGUGAGCAA___AACA .................................................(((((..((((((((((((...))))))))))))..))))).(((((((.....)))))))......... (-18.45 = -18.30 + -0.15)
| Location | 17,574,616 – 17,574,710 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Shannon entropy | 0.39973 |
| G+C content | 0.36534 |
| Mean single sequence MFE | -22.99 |
| Consensus MFE | -17.13 |
| Energy contribution | -16.97 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.997041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17574616 94 - 23011544 UGUU---UUGCUCACAG-UCGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGACCCUACACACCCAUACAUCCCC--------------------- .(((---((((....((-((((....))))))....(((((((((((.....))))))))))))))))))............................--------------------- ( -23.00, z-score = -4.57, R) >droSim1.chr2L 17277837 94 - 22036055 UGUU---UUGCUCACAG-UCGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCCGACCCUACACACCCAUACAUCCAC--------------------- .(((---((((....((-((((....))))))....(((((((((((.....))))))))))))))))))............................--------------------- ( -23.00, z-score = -4.37, R) >droSec1.super_7 1215454 94 - 3727775 UGUU---UUGCUCACAG-UCGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCCGACCCUACACACCCAUACAUCCAC--------------------- .(((---((((....((-((((....))))))....(((((((((((.....))))))))))))))))))............................--------------------- ( -23.00, z-score = -4.37, R) >droYak2.chr2R 17614005 91 + 21139217 UGUU---UUGCUCACAG-UCAUGCAAACGAUUUUCAACACAUUUACAAUUACUGUAAAUGUGUGCAAAAUUUCAGACC---CACACCCAUACAUCCAC--------------------- .(((---((((....((-((........))))....(((((((((((.....))))))))))))))))))........---.................--------------------- ( -18.70, z-score = -2.89, R) >droEre2.scaffold_4845 15912966 93 + 22589142 UGUU---UUGCUCGCAG-UCGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGAGCCCACACACCCAUACAUCCA---------------------- (((.---..((((..((-((((....))))))....(((((((((((.....)))))))))))...........))))..)))..............---------------------- ( -23.50, z-score = -3.68, R) >droAna3.scaffold_12916 12878296 91 + 16180835 UGCU---UUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGACCCACUACCAAUACACACA------------------------ ...(---((((....((-((((....))))))....(((((((((((.....))))))))))))))))...........................------------------------ ( -19.20, z-score = -2.21, R) >dp4.chr4_group3 833033 102 + 11692001 UGUU---UUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGGCCCCACACUCACACAUAAGACUGUCUCCGU------------- .(((---((((....((-((((....))))))....(((((((((((.....))))))))))))))))))....((....(((.((........)).)))..))..------------- ( -23.30, z-score = -1.99, R) >droPer1.super_8 1896219 102 + 3966273 UGUU---UUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGGCCCCACACUCACACAUAAGCAGGUCUCCGU------------- .(((---((((....((-((((....))))))....(((((((((((.....))))))))))))))))))...(((((...................)))))....------------- ( -24.01, z-score = -2.01, R) >droWil1.scaffold_180764 3020211 116 + 3949147 GAUU---UUGCUCACAGUUUGUGCAAACGAUUUUCAACACAUUUACAAUAAUUGUAAAUGUGUGCAAAAUUUCAGACUUUACAGCAAUACAUAUACAUAUUACGGAGAAGCUCUUUCCA ....---(((((...((((((.......((((((..(((((((((((.....)))))))))))..)))))).))))))....)))))................(((((.....))))). ( -26.40, z-score = -3.22, R) >droVir3.scaffold_12963 16502557 92 - 20206255 UGUUCGGUUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAAACAUUAUGCACGUUUGUG-------------------------- ............(((((-.((((((...((((((..(((((((((((.....)))))))))))..))))))..........)))))).)))))-------------------------- ( -23.62, z-score = -1.62, R) >droMoj3.scaffold_6500 21428995 91 - 32352404 UGUU-GGUUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAAACAUUAUGCGCGUGUGUG-------------------------- ....-.......((((.-.((((((...((((((..(((((((((((.....)))))))))))..))))))..........))))))..))))-------------------------- ( -22.32, z-score = -0.30, R) >droGri2.scaffold_15252 13917111 91 - 17193109 UGUU-GCUUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAAUAUUGUAAAUGUGUGCAAAAUUUCAAGCAUUAUGCAUGUGUGUG-------------------------- ....-....((.(((((-(.((((....((((((..(((((((((((.....)))))))))))..))))))....))))...)).)))).)).-------------------------- ( -25.80, z-score = -1.50, R) >consensus UGUU___UUGCUCACAG_UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGACCCUACACACCCAUACAUCCA______________________ .......((((.......((((....))))......(((((((((((.....))))))))))))))).................................................... (-17.13 = -16.97 + -0.16)
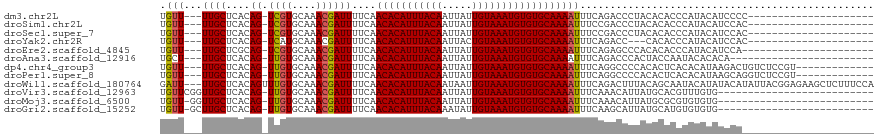
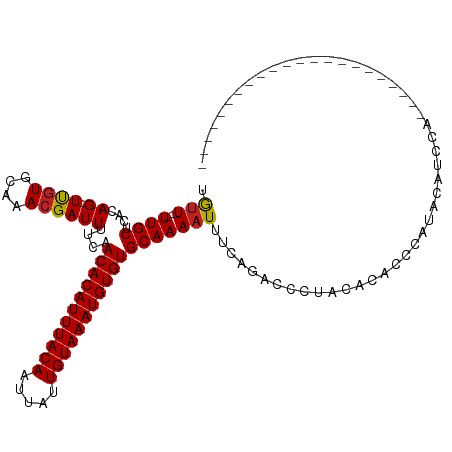
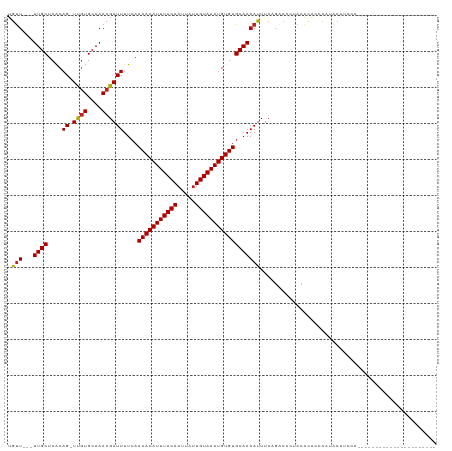
| Location | 17,574,634 – 17,574,739 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.57 |
| Shannon entropy | 0.41564 |
| G+C content | 0.36906 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -18.45 |
| Energy contribution | -18.30 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.967680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17574634 105 + 23011544 AGGGUCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACGA-CUGUGAGCAA-----AACACGUCGGAGAAGAAAUUACUCUCUCGCUGU------- ..........(((((..(((((((((((.....)))))))))))..))))).....((.(((-((((......-----.))).))))((((.((......))))))))...------- ( -26.10, z-score = -1.44, R) >droSim1.chr2L 17277855 105 + 22036055 AGGGUCGGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACGA-CUGUGAGCAA-----AACACGUCGGAGAAGAAAUUACUCUCUCGCUGU------- ...((..(..(((((..(((((((((((.....)))))))))))..)))))..)..)).(((-((((......-----.))).))))((((.((......)))))).....------- ( -26.60, z-score = -1.45, R) >droSec1.super_7 1215472 105 + 3727775 AGGGUCGGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACGA-CUGUGAGCAA-----AACACGUCGGAGAAGAAAUUACUCUCUCGCUGU------- ...((..(..(((((..(((((((((((.....)))))))))))..)))))..)..)).(((-((((......-----.))).))))((((.((......)))))).....------- ( -26.60, z-score = -1.45, R) >droYak2.chr2R 17614023 102 - 21139217 ---GUCUGAAAUUUUGCACACAUUUACAGUAAUUGUAAAUGUGUUGAAAAUCGUUUGCAUGA-CUGUGAGCAA-----AACACUUCGGAGAAGAAAUUGCUCUCUCGCUGU------- ---.(((((((((((..((((((((((((...))))))))))))..))))).(((..((...-.))..)))..-----.....))))))(((((......))).)).....------- ( -24.80, z-score = -1.16, R) >droEre2.scaffold_4845 15912983 105 - 22589142 GGGCUCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACGA-CUGCGAGCAA-----AACACGUCGGAGAAGAAAUUACUCUCUCGCUGU------- .((((((((..(((((((((((((((((.....)))))))))))..........(((((...-.)))))))))-----))....)))))(((((......))).)))))..------- ( -29.90, z-score = -2.75, R) >droAna3.scaffold_12916 12878312 104 - 16180835 -GGGUCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAA-----AGCACGUCGGAGUAGAAAUCACUCUCUUUGUGU------- -...........((((((((((((((((.....))))))))))).............(((..-..))).))))-----)(((((..((((((....).)))))...)))))------- ( -27.40, z-score = -1.72, R) >dp4.chr4_group3 833059 105 - 11692001 GGGGCCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAA-----AACACGUCGGAGUAGAAAUCACUCUCUCUCUCU------- ..(((.((..(((((..(((((((((((.....)))))))))))..))))).(((..((...-.))..)))..-----..)).)))((((.(((......)))))))....------- ( -26.90, z-score = -2.42, R) >droPer1.super_8 1896245 112 - 3966273 GGGGCCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAA-----AACACGUCGGAGUAGAAAUCACUCUCUCUCUCUGUGUGUG ............((((((((((((((((.....))))))))))).............(((..-..))).))))-----)(((((.(((((.(((........)))..)))))))))). ( -31.00, z-score = -2.40, R) >droWil1.scaffold_180764 3020250 110 - 3949147 AAAGUCUGAAAUUUUGCACACAUUUACAAUUAUUGUAAAUGUGUUGAAAAUCGUUUGCACAAACUGUGAGCAAAAUCCUGUAGCUGGGAGUGAAAAUAUCUCCAUGAUAA-------- ..((.(((..((((((((((((((((((.....))))))))))).............(((.....))).)))))))....))))).((((.........)))).......-------- ( -26.70, z-score = -2.34, R) >droVir3.scaffold_12963 16502570 90 + 20206255 AAUGUUUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAACCG--AACAGACAUGUGUGG------------------------- .(((((((...(((((.(((((((((((.....)))))))))))))))).(((.((((.((.-...)).)))).))--).)))))))......------------------------- ( -26.70, z-score = -2.72, R) >droMoj3.scaffold_6500 21429008 88 + 32352404 AAUGUUUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAACC---AACAG-CAUGUGUAU------------------------- ..........(((((..(((((((((((.....)))))))))))..)))))....((((((.-((((........---.))))-..)))))).------------------------- ( -21.50, z-score = -1.07, R) >droGri2.scaffold_15252 13917124 89 + 17193109 AAUGCUUGAAAUUUUGCACACAUUUACAAUAUUUGUAAAUGUGUUGAAAAUCGUUUGCACAA-CUGUGAGCAAGC---AACACACAUGUGUGU------------------------- ..(((((...(((((..(((((((((((.....)))))))))))..))))).(((..((...-.))..)))))))---)(((((....)))))------------------------- ( -27.80, z-score = -2.37, R) >consensus AGGGUCUGAAAUUUUGCACACAUUUACAAUAAUUGUAAAUGUGUUGAAAAUCGUUUGCACAA_CUGUGAGCAA_____AACACGUCGGAGUAGAAAUUACUCUCUCGCUGU_______ ..........(((((..((((((((((((...))))))))))))..))))).(((((((.....)))))))............................................... (-18.45 = -18.30 + -0.15)

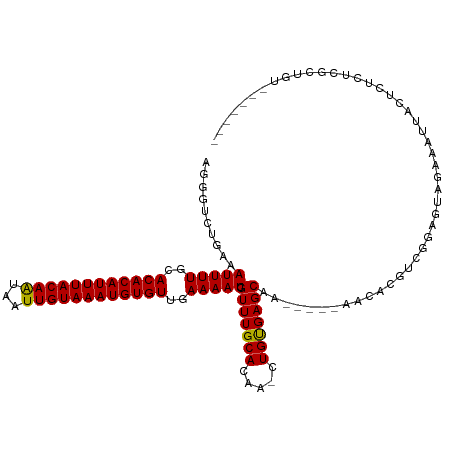
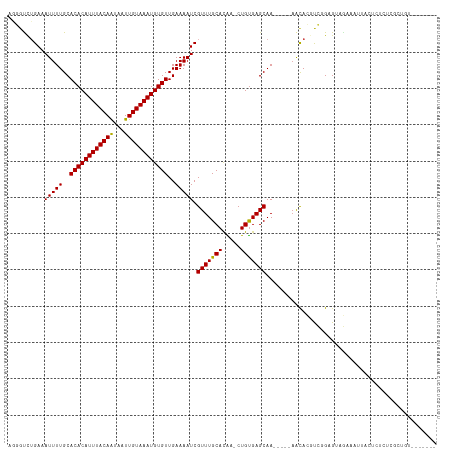
| Location | 17,574,634 – 17,574,739 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.57 |
| Shannon entropy | 0.41564 |
| G+C content | 0.36906 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -17.14 |
| Energy contribution | -16.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17574634 105 - 23011544 -------ACAGCGAGAGAGUAAUUUCUUCUCCGACGUGUU-----UUGCUCACAG-UCGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGACCCU -------...(.(((((((....))).)))))((...(((-----((((....((-((((....))))))....(((((((((((.....)))))))))))))))))).))....... ( -29.30, z-score = -3.56, R) >droSim1.chr2L 17277855 105 - 22036055 -------ACAGCGAGAGAGUAAUUUCUUCUCCGACGUGUU-----UUGCUCACAG-UCGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCCGACCCU -------...(.(((((((....))).)))))..((.(((-----((((....((-((((....))))))....(((((((((((.....))))))))))))))))))...))..... ( -29.60, z-score = -3.79, R) >droSec1.super_7 1215472 105 - 3727775 -------ACAGCGAGAGAGUAAUUUCUUCUCCGACGUGUU-----UUGCUCACAG-UCGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCCGACCCU -------...(.(((((((....))).)))))..((.(((-----((((....((-((((....))))))....(((((((((((.....))))))))))))))))))...))..... ( -29.60, z-score = -3.79, R) >droYak2.chr2R 17614023 102 + 21139217 -------ACAGCGAGAGAGCAAUUUCUUCUCCGAAGUGUU-----UUGCUCACAG-UCAUGCAAACGAUUUUCAACACAUUUACAAUUACUGUAAAUGUGUGCAAAAUUUCAGAC--- -------...((((((((((((...((((...))))....-----))))))...(-((........))))))).(((((((((((.....)))))))))))))............--- ( -27.20, z-score = -2.77, R) >droEre2.scaffold_4845 15912983 105 + 22589142 -------ACAGCGAGAGAGUAAUUUCUUCUCCGACGUGUU-----UUGCUCGCAG-UCGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGAGCCC -------.....((((((....))))))(((.((...(((-----((((....((-((((....))))))....(((((((((((.....)))))))))))))))))).)).)))... ( -31.40, z-score = -3.64, R) >droAna3.scaffold_12916 12878312 104 + 16180835 -------ACACAAAGAGAGUGAUUUCUACUCCGACGUGCU-----UUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGACCC- -------.(((...(.(((((.....))))))...))).(-----((((....((-((((....))))))....(((((((((((.....))))))))))))))))...........- ( -25.60, z-score = -2.33, R) >dp4.chr4_group3 833059 105 + 11692001 -------AGAGAGAGAGAGUGAUUUCUACUCCGACGUGUU-----UUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGGCCCC -------.(.((((((((....))))).))))((...(((-----((((....((-((((....))))))....(((((((((((.....)))))))))))))))))).))....... ( -30.00, z-score = -3.15, R) >droPer1.super_8 1896245 112 + 3966273 CACACACAGAGAGAGAGAGUGAUUUCUACUCCGACGUGUU-----UUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGGCCCC .((((...(.((((((((....))))).))))...))))(-----((((....((-((((....))))))....(((((((((((.....))))))))))))))))............ ( -30.80, z-score = -2.80, R) >droWil1.scaffold_180764 3020250 110 + 3949147 --------UUAUCAUGGAGAUAUUUUCACUCCCAGCUACAGGAUUUUGCUCACAGUUUGUGCAAACGAUUUUCAACACAUUUACAAUAAUUGUAAAUGUGUGCAAAAUUUCAGACUUU --------.......((((.........))))........(((((((((.....((((....))))........(((((((((((.....))))))))))))))))))))........ ( -24.70, z-score = -2.36, R) >droVir3.scaffold_12963 16502570 90 - 20206255 -------------------------CCACACAUGUCUGUU--CGGUUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAAACAUU -------------------------......((((.((..--...((((....((-((((....))))))....(((((((((((.....))))))))))))))).....)).)))). ( -22.20, z-score = -2.21, R) >droMoj3.scaffold_6500 21429008 88 - 32352404 -------------------------AUACACAUG-CUGUU---GGUUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAAACAUU -------------------------...((((.(-((((.---((...)).))))-))))).....((((((..(((((((((((.....)))))))))))..))))))......... ( -21.80, z-score = -1.74, R) >droGri2.scaffold_15252 13917124 89 - 17193109 -------------------------ACACACAUGUGUGUU---GCUUGCUCACAG-UUGUGCAAACGAUUUUCAACACAUUUACAAAUAUUGUAAAUGUGUGCAAAAUUUCAAGCAUU -------------------------(((((....)))))(---(((((..(((..-..))).....((((((..(((((((((((.....)))))))))))..)))))).)))))).. ( -26.00, z-score = -2.25, R) >consensus _______ACAGCGAGAGAGUAAUUUCUACUCCGACGUGUU_____UUGCUCACAG_UUGUGCAAACGAUUUUCAACACAUUUACAAUUAUUGUAAAUGUGUGCAAAAUUUCAGACCCU .............................................((((.......((((....))))......(((((((((((.....)))))))))))))))............. (-17.14 = -16.98 + -0.16)
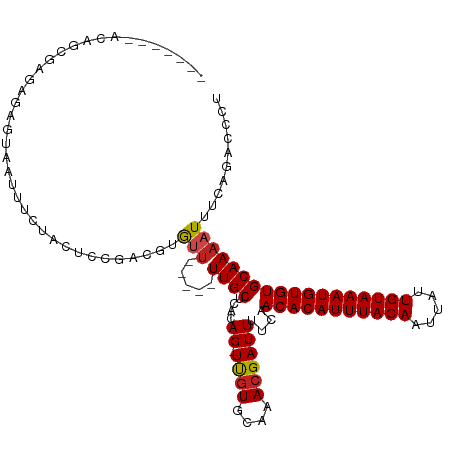
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:20 2011