
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,709,099 – 1,709,205 |
| Length | 106 |
| Max. P | 0.750480 |

| Location | 1,709,099 – 1,709,205 |
|---|---|
| Length | 106 |
| Sequences | 13 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 57.63 |
| Shannon entropy | 0.95847 |
| G+C content | 0.36755 |
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -7.73 |
| Energy contribution | -7.61 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.37 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.750480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1709099 106 + 23011544 UAGAACUGUGGGAAAAUAAACAUAUACAUGAUUAUGUAUAAACAAAGUCAUUUAGAAACACACUCGACCCCUACCGGCAGGAAUGGCCAAUCUAUGGCCGAGAUGA .....((((.((..........(((((((....)))))))......(((................))).....)).))))...((((((.....))))))...... ( -22.99, z-score = -1.46, R) >droSim1.chr2L 1719459 104 + 22036055 UAGAACUGUGGGAAAAUAAACAUACGUACA--UAAGCAUAAACAAAGUCAUUUAGAAACACACUCGUCCCCUACCGGCAGGAAUGGCCAAUCAAUGGCCGAGAUGU .....((((.((.............((...--...)).................((.......))........)).))))...((((((.....))))))...... ( -17.10, z-score = 0.66, R) >droSec1.super_14 1643699 104 + 2068291 UAGAACUGUGGGAAAAUAAACAUACGUACA--UAAGCAUAAACAAAGUCAUUUAGAAACACACUCGUCCCCUACCGGCAGGAAUGGCCAAUCUAUGGCCGAGAUGU .....((((.((.............((...--...)).................((.......))........)).))))...((((((.....))))))...... ( -17.10, z-score = 0.76, R) >droYak2.chr2L 1679845 101 + 22324452 UAGAACUGUGGGUAAAUAAACAGACAUAUG---AAGUAUAAACAAAUACAAUUUGAAGUA--CUCGUCGCCUACCGGCAGGAAUGGCCAAUCUAUGGCCGAGAUGU .....((((.((((........(((.....---..((((......)))).....((....--.)))))...)))).))))...((((((.....))))))...... ( -23.00, z-score = -1.25, R) >droEre2.scaffold_4929 1752793 101 + 26641161 UAGAACUGUGGGUAAAUAAGCAGGCAUAUG---AAACGUAAACAUAUUGAUUUUCAAGUA--CUCUUCGCCUACCGGCAGGAAUGGCCAGUCUAUGGCCGAGAUGU .....((((.(((........((((.((((---.........))))(((.....)))...--......))))))).))))...((((((.....))))))...... ( -24.80, z-score = -0.51, R) >droAna3.scaffold_12943 1063116 89 - 5039921 UAGAACUGU-AAACAAAAACCAUUAAUA-----UAGAAAUUCGAAUAUU----AGUCAACCACUU-------ACUGGUAGGAAUGGCCAGUCUAUGGCCGAUAUAA ......(((-(........(((((((((-----(.(.....)..)))))----)))..((((...-------..)))).))..((((((.....)))))).)))). ( -16.90, z-score = -1.46, R) >dp4.chr4_group3 8588286 98 - 11692001 UAGAACUA--ACAAACAAGUCAA-AUUAUGUAUAAGCU-AAACAGAACAAAUGAAGUGAUAAUAUCUU----ACUGGUUUGAAUGGCCAGUCUAUGGCUGAGAUAU ........--........((((.-.((((.......((-....)).....))))..)))).(((((((----(((((((.....)))))))........))))))) ( -14.20, z-score = 0.24, R) >droPer1.super_1 5696342 98 - 10282868 UAGAACUA--ACAAACAAGUCAA-AUUAUGUAUAAGCU-AAACAAAACAAAUGAAGUGAUAAUAUCUU----ACUGGUACGAAUGGCCAGUCUAUGGCUGAGAUAU ........--........((((.-.((((.........-...........))))..)))).(((((((----((((((.......))))))........))))))) ( -12.75, z-score = 0.27, R) >droWil1.scaffold_180708 3603795 103 + 12563649 UAGAACUG--AAACAGAAAUAAAUGACAAUUAAUUGAUGUACGAAUAUUUAUAAGUAAAUUGUUUCUUUU-UACCGGUAGAAACGGCCAAUCAAUGGCCGAUAUAA ....((((--(((.((((((((...((..(((...(((((....)))))..)))))...)))))))).))-)..)))).....((((((.....))))))...... ( -18.90, z-score = -1.87, R) >droVir3.scaffold_12963 7811794 101 + 20206255 UAGAACUGC-AAAAGAUAAUUUC-AAUUUAUAUUGUAUAUUUUAACAGAUGAGACUCGAAAAUUGUUUU---ACCGGCAAGAAUGGCCAAUCGAUGGCUGAUAUAU ......(((-..(((((((((((-..(((((.((((........)))))))))....).))))))))))---....))).....(((((.....)))))....... ( -14.50, z-score = 0.22, R) >droMoj3.scaffold_6500 29313331 99 - 32352404 UAGAACUGU-AAAAGUUAGUUAA-AAGAUAUAGUAAC--AUAUAGCAGAAAUAACUUGCUCAAAUUUAU---ACCGGCAAAAAGGGCCAAUCGAUGGCUGAUAUGU .........-....((((((((.-..(((...((((.--....(((((.......))))).....))))---...(((.......))).)))..)))))))).... ( -15.00, z-score = -0.02, R) >droGri2.scaffold_15252 10896253 98 + 17193109 UAGAACUGC-AAAAAAUUAUGAC-AGCGU-UAACAACAGAUAUAUAAAGGGCUACUCGUCAUGAUUUA-----CCGGCAAGAAUGGCCAAUCUAUAGCCGAUAUAU ......(((-...((((((((((-(((.(-(................)).)))....)))))))))).-----...)))....((((.........))))...... ( -16.39, z-score = -0.38, R) >anoGam1.chr3R 3143414 101 + 53272125 UAAAACUGU---GAAUUGGAUAG-AGUAGGGCAUGAAUAUGUCAGUCGUAAUAAUUCACCC-CACCACACUGACCGGUAAAAGCGGCCAAUCAACGGCUGACAAAU .......((---((((((....(-(....(((((....)))))..))....))))))))..-.(((.........)))....((((((.......))))).).... ( -20.60, z-score = 0.05, R) >consensus UAGAACUGU_AAAAAAUAAUCAA_AAUAUGUAUAAGCAUAAACAAAAUAAAUUAGUAGAACACUCCUC____ACCGGCAGGAAUGGCCAAUCUAUGGCCGAGAUAU ...................................................................................((((((.....))))))...... ( -7.73 = -7.61 + -0.12)

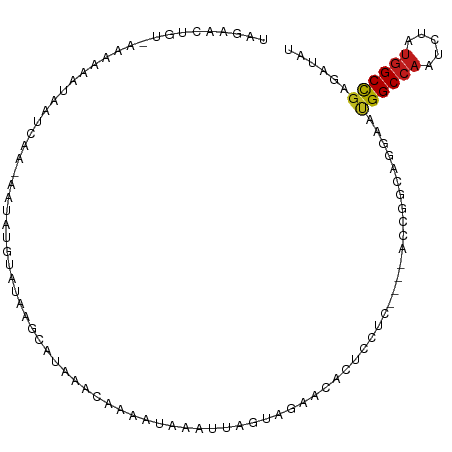
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:59 2011