
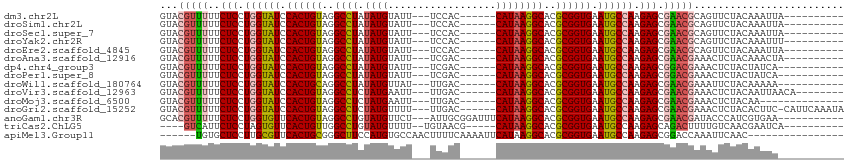
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,566,376 – 17,566,471 |
| Length | 95 |
| Max. P | 0.999179 |

| Location | 17,566,376 – 17,566,471 |
|---|---|
| Length | 95 |
| Sequences | 15 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.68 |
| Shannon entropy | 0.38201 |
| G+C content | 0.45846 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -28.57 |
| Energy contribution | -28.52 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
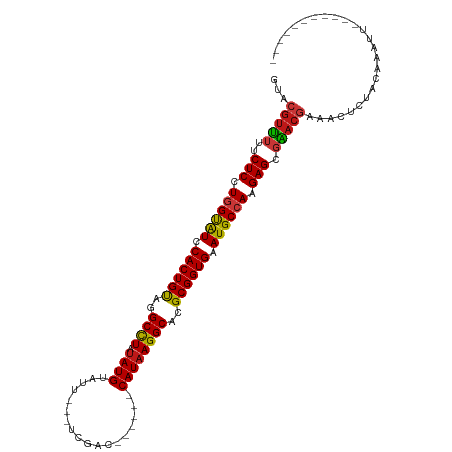
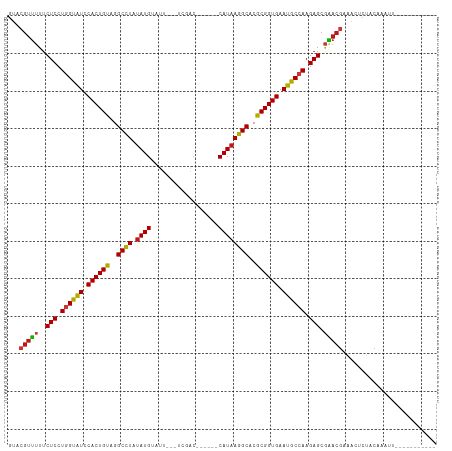
>dm3.chr2L 17566376 95 + 23011544 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUAUAUGUAUU---UCCAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGCAGUUCUACAAAUUA---------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))...............---------- ( -31.30, z-score = -3.63, R) >droSim1.chr2L 17270511 95 + 22036055 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUAUAUGUAUU---UCCAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGCAGUUCUACAAAUUA---------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))...............---------- ( -31.30, z-score = -3.63, R) >droSec1.super_7 1207326 95 + 3727775 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUAUAUGUAUU---UCCAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGCAGUUCUACAAAUUA---------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))...............---------- ( -31.30, z-score = -3.63, R) >droYak2.chr2R 17605945 95 - 21139217 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUAUAUGUAUU---UCCAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGCAGUUCUACAAAUUU---------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))...............---------- ( -31.30, z-score = -3.75, R) >droEre2.scaffold_4845 15905082 95 - 22589142 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUAUAUGUAUU---UCCAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGCAGUUCUACAAAUUA---------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))...............---------- ( -31.30, z-score = -3.63, R) >droAna3.scaffold_12916 12871051 95 - 16180835 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUAUAUGUAUU---UCGAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGAAACUCUACAAACUA---------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))...............---------- ( -32.00, z-score = -4.04, R) >dp4.chr4_group3 824283 94 - 11692001 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUAUAUGUAUU---UCGAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGGACGAAACUCUACUAUCA----------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))..............----------- ( -31.60, z-score = -3.32, R) >droPer1.super_8 1887458 94 - 3966273 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUAUAUGUAUU---UCGAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGGACGAAACUCUACUAUCA----------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))..............----------- ( -31.60, z-score = -3.32, R) >droWil1.scaffold_180764 3008858 94 - 3949147 GUACGUUUUUCUCCUGGUAUCCACUGCAGGCCUAUAUGUUAU---UUGAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGAAAUUCUACAAAAA----------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))..............----------- ( -33.30, z-score = -4.63, R) >droVir3.scaffold_12963 16493169 97 + 20206255 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUCUAUGAAUU---UUGAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGAAACUCUACAAUUAACA-------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).))))).................-------- ( -32.50, z-score = -4.34, R) >droMoj3.scaffold_6500 21419493 91 + 32352404 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUCUAUGAAUU---UUGAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGAAACUCUACAA-------------- ...(((((..(((.((((((.((((((..((((.((((....---.....------))))))))..)))))).)))))).))).)))))...........-------------- ( -32.50, z-score = -4.48, R) >droGri2.scaffold_15252 13906834 104 + 17193109 GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUCUAUGUUUU---UUGAC------CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGAAACUCUACACUUC-CAUUCAAAUA ...(((((..(((.((((((.((((((..((((.((((((..---..)).------))))))))..)))))).)))))).))).)))))..............-.......... ( -32.30, z-score = -3.89, R) >anoGam1.chr3R 29342229 100 + 53272125 GCACGUUUUUCUCCUGGUGUUCACUGUAGGCCUGUAUGUUCU---AUUGCGGAUUUCAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGAUACCCAUCGUGAA----------- .(((((((..(((.(((((((((((((..((((.((((.(((---.....)))...))))))))..))))))))))))).))).))))(((....))))))..----------- ( -39.20, z-score = -3.93, R) >triCas2.ChLG5 1908647 93 - 18847211 ----GUCAUUCUCCUAGUGUUCACUGUUGGCCUGUAUGUUUU--UGUAACG-----CAUAAGGCACGCGGUGAAUGCCAAGAGCAGACUUUUGUCAACGAAUCA---------- ----.((...(((...(((((((((((..((((.(((((...--......)-----))))))))..)))))))))))...)))..(((....)))...))....---------- ( -28.00, z-score = -1.85, R) >apiMel3.Group11 8130434 92 - 12576330 ------UGUGCUCCUUGCGUUCACUGCGGGCUUCCAUGUGCCAACUUUUCAAAAUUCAUAAGGCACGCGGUGAAUGCCAAGAGCGGACCAAAUUCAAC---------------- ------..(((((...(((((((((((((....))..(((((...................))))))))))))))))...))))).............---------------- ( -33.81, z-score = -3.37, R) >consensus GUACGUUUUUCUCCUGGUAUCCACUGUAGGCCUAUAUGUAUU___UCGAC______CAUAAGGCACGCGGUGAAUGCCAAGAGCGAACGAAACUCUACAAAUU___________ ...(((((..(((.((((((.((((((..((((.((((..................))))))))..)))))).)))))).))).)))))......................... (-28.57 = -28.52 + -0.06)
| Location | 17,566,376 – 17,566,471 |
|---|---|
| Length | 95 |
| Sequences | 15 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.68 |
| Shannon entropy | 0.38201 |
| G+C content | 0.45846 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -21.39 |
| Energy contribution | -21.61 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
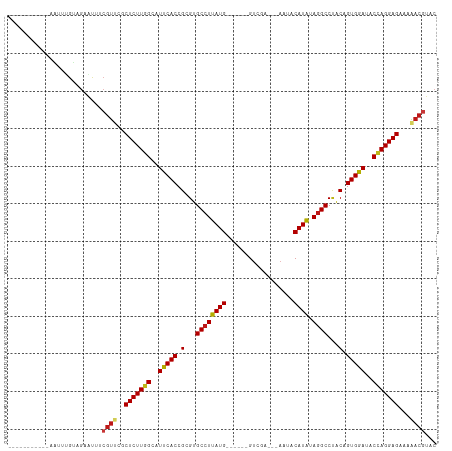
>dm3.chr2L 17566376 95 - 23011544 ----------UAAUUUGUAGAACUGCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUGGA---AAUACAUAUAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC ----------.............((((((..(((((((.((((((.(...((((((((------((...---.)).)))).))))...).)))))).)))))))...)))))). ( -31.30, z-score = -2.99, R) >droSim1.chr2L 17270511 95 - 22036055 ----------UAAUUUGUAGAACUGCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUGGA---AAUACAUAUAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC ----------.............((((((..(((((((.((((((.(...((((((((------((...---.)).)))).))))...).)))))).)))))))...)))))). ( -31.30, z-score = -2.99, R) >droSec1.super_7 1207326 95 - 3727775 ----------UAAUUUGUAGAACUGCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUGGA---AAUACAUAUAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC ----------.............((((((..(((((((.((((((.(...((((((((------((...---.)).)))).))))...).)))))).)))))))...)))))). ( -31.30, z-score = -2.99, R) >droYak2.chr2R 17605945 95 + 21139217 ----------AAAUUUGUAGAACUGCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUGGA---AAUACAUAUAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC ----------.............((((((..(((((((.((((((.(...((((((((------((...---.)).)))).))))...).)))))).)))))))...)))))). ( -31.30, z-score = -3.03, R) >droEre2.scaffold_4845 15905082 95 + 22589142 ----------UAAUUUGUAGAACUGCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUGGA---AAUACAUAUAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC ----------.............((((((..(((((((.((((((.(...((((((((------((...---.)).)))).))))...).)))))).)))))))...)))))). ( -31.30, z-score = -2.99, R) >droAna3.scaffold_12916 12871051 95 + 16180835 ----------UAGUUUGUAGAGUUUCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUCGA---AAUACAUAUAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC ----------...............((((..(((((((.((((((.(...((((((((------.....---....)))).))))...).)))))).)))))))...))))... ( -28.00, z-score = -2.02, R) >dp4.chr4_group3 824283 94 + 11692001 -----------UGAUAGUAGAGUUUCGUCCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUCGA---AAUACAUAUAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC -----------..............(((...(((((((.((((((.(...((((((((------.....---....)))).))))...).)))))).)))))))....)))... ( -26.70, z-score = -1.57, R) >droPer1.super_8 1887458 94 + 3966273 -----------UGAUAGUAGAGUUUCGUCCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUCGA---AAUACAUAUAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC -----------..............(((...(((((((.((((((.(...((((((((------.....---....)))).))))...).)))))).)))))))....)))... ( -26.70, z-score = -1.57, R) >droWil1.scaffold_180764 3008858 94 + 3949147 -----------UUUUUGUAGAAUUUCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUCAA---AUAACAUAUAGGCCUGCAGUGGAUACCAGGAGAAAAACGUAC -----------..............((((..(((((((.((((((.(((.((((((((------.....---....)))).)))).))).)))))).)))))))...))))... ( -33.70, z-score = -4.35, R) >droVir3.scaffold_12963 16493169 97 - 20206255 --------UGUUAAUUGUAGAGUUUCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUCAA---AAUUCAUAGAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC --------.................((((..(((((((.((((((.(...((((((((------(....---...)))).)))))...).)))))).)))))))...))))... ( -28.80, z-score = -2.34, R) >droMoj3.scaffold_6500 21419493 91 - 32352404 --------------UUGUAGAGUUUCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUCAA---AAUUCAUAGAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC --------------...........((((..(((((((.((((((.(...((((((((------(....---...)))).)))))...).)))))).)))))))...))))... ( -28.80, z-score = -2.73, R) >droGri2.scaffold_15252 13906834 104 - 17193109 UAUUUGAAUG-GAAGUGUAGAGUUUCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG------GUCAA---AAAACAUAGAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC .....(((((-(((.(....).)))))))).(((((((.((((((.(...((((((((------.....---....))).)))))...).)))))).))))))).......... ( -32.10, z-score = -2.68, R) >anoGam1.chr3R 29342229 100 - 53272125 -----------UUCACGAUGGGUAUCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUGAAAUCCGCAAU---AGAACAUACAGGCCUACAGUGAACACCAGGAGAAAAACGUGC -----------..((((((....)))(((..(((((((..(((((.(...((((((((...........---....)))).))))...).)))))..)))))))...)))))). ( -26.26, z-score = -0.91, R) >triCas2.ChLG5 1908647 93 + 18847211 ----------UGAUUCGUUGACAAAAGUCUGCUCUUGGCAUUCACCGCGUGCCUUAUG-----CGUUACA--AAAACAUACAGGCCAACAGUGAACACUAGGAGAAUGAC---- ----------....((((((((....)))..(((((((..(((((.(...((((((((-----.......--....)))).))))...).)))))..)))))))))))).---- ( -24.10, z-score = -1.44, R) >apiMel3.Group11 8130434 92 + 12576330 ----------------GUUGAAUUUGGUCCGCUCUUGGCAUUCACCGCGUGCCUUAUGAAUUUUGAAAAGUUGGCACAUGGAAGCCCGCAGUGAACGCAAGGAGCACA------ ----------------..............((((((.((.(((((.(((((((...................)))))..((....)))).))))).)).))))))...------ ( -30.51, z-score = -1.43, R) >consensus ___________AAUUUGUAGAAUUUCGUUCGCUCUUGGCAUUCACCGCGUGCCUUAUG______GUCGA___AAUACAUAUAGGCCUACAGUGGAUACCAGGAGAAAAACGUAC .........................((((..(((((((..(((((.(...((((((((..................)))).))))...).)))))..)))))))...))))... (-21.39 = -21.61 + 0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:16 2011