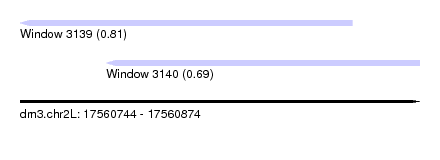
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,560,744 – 17,560,874 |
| Length | 130 |
| Max. P | 0.814603 |

| Location | 17,560,744 – 17,560,852 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 92.53 |
| Shannon entropy | 0.15645 |
| G+C content | 0.47608 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -23.39 |
| Energy contribution | -23.49 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
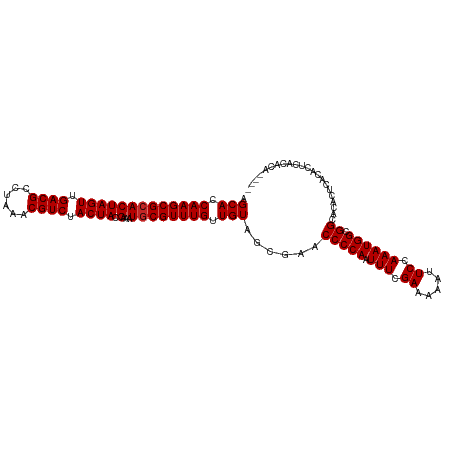
>dm3.chr2L 17560744 108 - 23011544 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGCACAUUCGCACUCACACU---- .(((.(((((((((((((.((((......)))).)))).)...)))))))).))).((((((((((.(((.((.....)).)))))).))....))))).........---- ( -30.00, z-score = -2.80, R) >droSim1.chr2L 17264834 108 - 22036055 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGCACACUCGCACUCACACC---- .(((.(((((((((((((.((((......)))).)))).)...)))))))).))).((((.(((((.(((.((.....)).)))))).)).....)))).........---- ( -28.50, z-score = -2.32, R) >droSec1.super_7 1201625 108 - 3727775 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUACGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGCACACUCGCACUCACACC---- .(((.((((((...((((.((((......)))).)))).......)))))).))).((((.(((((.(((.((.....)).)))))).)).....)))).........---- ( -23.20, z-score = -1.12, R) >droYak2.chr2R 17600400 108 + 21139217 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGCACACUCAAACUCAUACC---- .(((.(((((((((((((.((((......)))).)))).)...)))))))).)))((.(..(((((.(((.((.....)).)))))).))..).))............---- ( -24.20, z-score = -1.56, R) >droEre2.scaffold_4845 15899579 108 + 22589142 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGCACACUCGCACUCACACC---- .(((.(((((((((((((.((((......)))).)))).)...)))))))).))).((((.(((((.(((.((.....)).)))))).)).....)))).........---- ( -28.50, z-score = -2.32, R) >dp4.chr4_group3 817289 112 + 11692001 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGUACACACACACACGUGCAGCCA .(((((((((((((((((.((((......)))).)))).)...)))))))).(((.(.(.((((((.(((.((.....)).)))))).))).).)))).....))))..... ( -30.40, z-score = -1.82, R) >droPer1.super_8 1880565 110 + 3966273 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGUACACACACACAUGCAUACA-- .(((.(((((((((((((.((((......)))).)))).)...)))))))).(((.(.(.((((((.(((.((.....)).)))))).))).).))))....))).....-- ( -27.40, z-score = -1.87, R) >droVir3.scaffold_12963 16486126 108 - 20206255 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAUUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGCACACGCACACAAGUGUA---- .(((((((((((((((((.((((......)))).)))).)...)))))))).(((.(((..(((((.(((.((.....)).)))))).))....))))))...)))).---- ( -29.30, z-score = -1.52, R) >droMoj3.scaffold_6500 21412186 108 - 32352404 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAUUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGCACACGCAUACAGCUGCA---- .(((..((((((((((((.((((......)))).)))).)...)))))))(((((((((..(((((.(((.((.....)).)))))).))....))).))))))))).---- ( -31.40, z-score = -2.07, R) >droGri2.scaffold_15252 13899159 108 - 17193109 AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAUUGCGUUUGUUGUAGCAAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGCACACGCACACUACGACA---- ......((((((((((((.((((......)))).)))).)...)))))))(((((((.........((((.((.....)).)))).(((.....)))...))))))).---- ( -26.40, z-score = -1.45, R) >consensus AGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUCCAAAUGGCGGCACACUCACACUCACACA____ .(((.(((((((((((((.((((......)))).)))).)...)))))))).)))......(((((.(((.((.....)).)))))).))...................... (-23.39 = -23.49 + 0.10)


| Location | 17,560,772 – 17,560,874 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Shannon entropy | 0.19554 |
| G+C content | 0.47366 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -24.33 |
| Energy contribution | -24.49 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.685643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17560772 102 - 23011544 -----------AGCGUGCUU-UAC-GUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC -----------.((((((((-...-....)))))).(((.(((((((((((((.((((......)))).)))).)...)))))))).))).))...................... ( -29.60, z-score = -1.89, R) >droSim1.chr2L 17264862 102 - 22036055 -----------AGCGUGCUU-UAC-GUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC -----------.((((((((-...-....)))))).(((.(((((((((((((.((((......)))).)))).)...)))))))).))).))...................... ( -29.60, z-score = -1.89, R) >droSec1.super_7 1201653 102 - 3727775 -----------AGCGUGCUU-UAC-GUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUACGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC -----------.((((((((-...-....)))))).((.....))))..((((.((((......)))).))))((((((..((((((....))))))....))))))........ ( -27.30, z-score = -1.58, R) >droYak2.chr2R 17600428 102 + 21139217 -----------AGCGUGCUU-UAC-GUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC -----------.((((((((-...-....)))))).(((.(((((((((((((.((((......)))).)))).)...)))))))).))).))...................... ( -29.60, z-score = -1.89, R) >droEre2.scaffold_4845 15899607 102 + 22589142 -----------AGCGUGCUU-UAC-GUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC -----------.((((((((-...-....)))))).(((.(((((((((((((.((((......)))).)))).)...)))))))).))).))...................... ( -29.60, z-score = -1.89, R) >droAna3.scaffold_12916 12865994 103 + 16180835 ---------UGAGCGAG-UU-UAC-GGGAAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC ---------....((((-..-...-(((....(((.(((.(((((((((((((.((((......)))).)))).)...)))))))).))).)))..)))....))))........ ( -32.50, z-score = -2.67, R) >dp4.chr4_group3 817321 103 + 11692001 ---------AGCGCGUG-UC-UAC-GGGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC ---------.((((.(.-.(-...-.)..).)))).(((.(((((((((((((.((((......)))).)))).)...)))))))).)))..((((.......))))........ ( -32.00, z-score = -1.72, R) >droPer1.super_8 1880595 103 + 3966273 ---------AGCGCGUG-UC-UAC-GGGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC ---------.((((.(.-.(-...-.)..).)))).(((.(((((((((((((.((((......)))).)))).)...)))))))).)))..((((.......))))........ ( -32.00, z-score = -1.72, R) >droWil1.scaffold_180764 3002221 114 + 3949147 CGAGUAGUGUGUGUAGAUUGAAAU-GUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAUUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC .................(((((((-.......(((.(((.(((((((((((((.((((......)))).)))).)...)))))))).))).))).......)))))))....... ( -31.24, z-score = -1.83, R) >droVir3.scaffold_12963 16486154 102 - 20206255 ------------GCGUGGUUACGU-UUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAUUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC ------------(((((.((((..-..)))).))).(((.(((((((((((((.((((......)))).)))).)...)))))))).))).))...................... ( -29.00, z-score = -1.48, R) >droMoj3.scaffold_6500 21412214 103 - 32352404 -----------AGCGUGGUUACGU-GUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAUUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC -----------...(.((((.(((-(((....))).(((.(((((((((((((.((((......)))).)))).)...)))))))).))).))))))).)............... ( -29.30, z-score = -1.11, R) >droGri2.scaffold_15252 13899187 106 - 17193109 ---------AACGUGGCUGAGCGUGGUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAUUGCGUUUGUUGUAGCAAACCCCAAUUUCGAAAAAUUC ---------...((((((..(.(((.(((....))).)))).))).)))((((.((((......)))).))))(((((((.((((((....))))))..))).))))........ ( -29.10, z-score = -0.56, R) >consensus ___________AGCGUGCUU_UAC_GUGUAAGCGCAGCACCAAGCGCACUAGUUGACGCCUAAACGUCUACUACGAAAUGCGUUUGUUGUAGCGAACCCCAAUUUCGAAAAAUUC ................................(((.(((.(((((((((((((.((((......)))).)))).)...)))))))).))).)))..................... (-24.33 = -24.49 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:14 2011