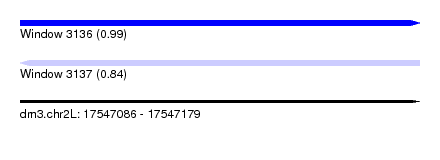
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,547,086 – 17,547,179 |
| Length | 93 |
| Max. P | 0.988229 |

| Location | 17,547,086 – 17,547,179 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 72.27 |
| Shannon entropy | 0.54568 |
| G+C content | 0.40051 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -14.89 |
| Energy contribution | -13.90 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
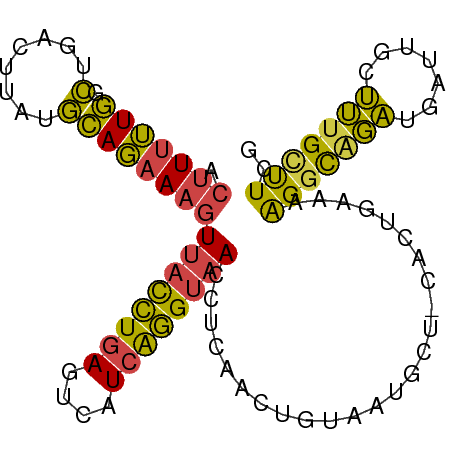
>dm3.chr2L 17547086 93 + 23011544 ACUUUUUGGCUGACUUAUGCAGAAAGUUACCUGAGUCAUCAGGUAACCUCAACUGUGAUGCU-CACUGAAAAGGCAGAUGAUUGCUUUGAUUCG .(((((..(.(((.....((((...(((((((((....))))))))).....)))).....)-)))..)))))((((....))))......... ( -26.00, z-score = -1.94, R) >droSim1.chr2L 17251237 93 + 22036055 ACUUUUUGGCUGACUUAUGCAGAAAGUUACCUGAGUCAUCAGGUAACCUCAACUGUAAUGCU-CACUGAAAAGGCAGAUGAUUGCUUUGCUUCG .(((((..(.(((....(((((...(((((((((....))))))))).....)))))....)-)))..)))))(((((.......))))).... ( -28.70, z-score = -2.87, R) >droSec1.super_7 1188517 93 + 3727775 ACUUUUUGGCUGACUUAUGCAGAAAGUUACCUGAGUCAUCAGGUAACCUCAACUGUAAUGCU-CACUGAAAAGGCAGAUGAUUGCUUUGCUUCG .(((((..(.(((....(((((...(((((((((....))))))))).....)))))....)-)))..)))))(((((.......))))).... ( -28.70, z-score = -2.87, R) >droYak2.chr2R 17588153 92 - 21139217 ACUUUUUGGCUGACUUUUGCAGAAAGUUACCUGACUCAUCAGGUAAGCUCAACGGCAUUGCU-CGCCGAAAAGGUUGAUGAUGGCUUUGCUUC- .(((((((((.((.(..(((....((((((((((....)))))).)))).....)))..).)-))))))))))....................- ( -26.80, z-score = -1.42, R) >droEre2.scaffold_4845 15888520 79 - 22589142 ACUUUUUGGCUGACUUUUGCAGAAAGUUACCUGACUCAUCAGGUAACCUCAAC-----UGUU-C-------AGGUGGAUGAUGGUUUUGCCU-- .......(((.((((...((((...(((((((((....))))))))).....)-----)))(-(-------(......))).))))..))).-- ( -21.40, z-score = -1.14, R) >droWil1.scaffold_180764 958502 93 + 3949147 AAGUAGUUUUUGA-UUAAAAAUAAAAUCAUUUUAGAAAUUGAGAAAAAACUAUAGGAACAAUAUUCUGUAAGGGCAUGCCAUAGUCAUGUUUAG ..(((((((((..-((((...((((.....))))....))))..))))))))).((((.....))))....((((((((....).))))))).. ( -13.30, z-score = -0.50, R) >consensus ACUUUUUGGCUGACUUAUGCAGAAAGUUACCUGAGUCAUCAGGUAACCUCAACUGUAAUGCU_CACUGAAAAGGCAGAUGAUUGCUUUGCUUCG .(((((((.(........))))))))((((((((....)))))))).........................(((((((.......))))))).. (-14.89 = -13.90 + -0.99)

| Location | 17,547,086 – 17,547,179 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 72.27 |
| Shannon entropy | 0.54568 |
| G+C content | 0.40051 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -10.94 |
| Energy contribution | -11.75 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17547086 93 - 23011544 CGAAUCAAAGCAAUCAUCUGCCUUUUCAGUG-AGCAUCACAGUUGAGGUUACCUGAUGACUCAGGUAACUUUCUGCAUAAGUCAGCCAAAAAGU .........(((......)))(((((..((.-.((....(((..((((((((((((....))))))))))))))).....))..))..))))). ( -22.70, z-score = -1.44, R) >droSim1.chr2L 17251237 93 - 22036055 CGAAGCAAAGCAAUCAUCUGCCUUUUCAGUG-AGCAUUACAGUUGAGGUUACCUGAUGACUCAGGUAACUUUCUGCAUAAGUCAGCCAAAAAGU ....((...((..(((.(((......)))))-)))....(((..((((((((((((....))))))))))))))).........))........ ( -22.90, z-score = -1.16, R) >droSec1.super_7 1188517 93 - 3727775 CGAAGCAAAGCAAUCAUCUGCCUUUUCAGUG-AGCAUUACAGUUGAGGUUACCUGAUGACUCAGGUAACUUUCUGCAUAAGUCAGCCAAAAAGU ....((...((..(((.(((......)))))-)))....(((..((((((((((((....))))))))))))))).........))........ ( -22.90, z-score = -1.16, R) >droYak2.chr2R 17588153 92 + 21139217 -GAAGCAAAGCCAUCAUCAACCUUUUCGGCG-AGCAAUGCCGUUGAGCUUACCUGAUGAGUCAGGUAACUUUCUGCAAAAGUCAGCCAAAAAGU -....................(((((.(((.-.((..(((.(..(((.((((((((....))))))))))).).)))...))..))).))))). ( -24.40, z-score = -1.64, R) >droEre2.scaffold_4845 15888520 79 + 22589142 --AGGCAAAACCAUCAUCCACCU-------G-AACA-----GUUGAGGUUACCUGAUGAGUCAGGUAACUUUCUGCAAAAGUCAGCCAAAAAGU --.(((.......(((......)-------)-).((-----(..((((((((((((....))))))))))))))).........)))....... ( -23.20, z-score = -2.96, R) >droWil1.scaffold_180764 958502 93 - 3949147 CUAAACAUGACUAUGGCAUGCCCUUACAGAAUAUUGUUCCUAUAGUUUUUUCUCAAUUUCUAAAAUGAUUUUAUUUUUAA-UCAAAAACUACUU ........((((((((...((..............))..))))))))..................(((((........))-))).......... ( -7.34, z-score = 0.40, R) >consensus CGAAGCAAAGCAAUCAUCUGCCUUUUCAGUG_AGCAUUACAGUUGAGGUUACCUGAUGACUCAGGUAACUUUCUGCAUAAGUCAGCCAAAAAGU .........................................((.((((((((((((....))))))))))))..)).................. (-10.94 = -11.75 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:11 2011