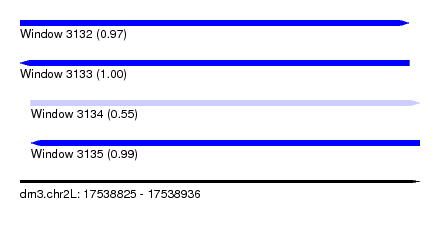
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,538,825 – 17,538,936 |
| Length | 111 |
| Max. P | 0.998583 |

| Location | 17,538,825 – 17,538,933 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 94.35 |
| Shannon entropy | 0.10190 |
| G+C content | 0.35196 |
| Mean single sequence MFE | -24.37 |
| Consensus MFE | -22.24 |
| Energy contribution | -23.28 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17538825 108 + 23011544 CAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCUUGACUUGGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCAGUUCCCACCUAUUGAACCCAUUUGUA ((((((((((((.(((......(((((...)))))..(((((((((....((.......))...))))))))).............))).)))))....))))))).. ( -22.00, z-score = -1.41, R) >droEre2.scaffold_4845 15880465 102 - 22589142 CAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCACUUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCGGUUCCCACCUAUUCAAACCA------ ......((((((.(((...(((((((.......((..(((((((((....((.......))...)))))))))..)).))))))).))).))))))......------ ( -23.60, z-score = -2.91, R) >droYak2.chr2R 17579705 108 - 21139217 CAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCACUUGACUUAGUCAACCGAAAAUGCCGAAAAUGAAGUCAUAUGUUUCGGUUCCCACCUAUUGAACCCAUUUGCA ((((((((((((.(((...............(((..(((((...)))))..)))...((((((((((....)))...)))))))..))).)))))....))))))).. ( -24.60, z-score = -2.28, R) >droSec1.super_7 1180450 108 + 3727775 CAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCGUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUAUUUCGGUUCCCACCUAUUGAACCCAUUUGUA ((((((((((((.(((...(((((((..........((((((((((....((.......))...))))))))))....))))))).))).)))))....))))))).. ( -25.14, z-score = -2.54, R) >droSim1.chr2L 17242876 108 + 22036055 CAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCUUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCGGUUCCCACCUAUUGAACCCAUUUGUA ((((((((((((.(((...............((((.(((((...))))).))))...(((((((..((.....))..)))))))..))).)))))....))))))).. ( -26.50, z-score = -3.02, R) >consensus CAAAUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCUUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCGGUUCCCACCUAUUGAACCCAUUUGUA ((((((((((((.(((...(((((((.......((..(((((((((....((.......))...)))))))))..)).))))))).))).)))))....))))))).. (-22.24 = -23.28 + 1.04)


| Location | 17,538,825 – 17,538,933 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.35 |
| Shannon entropy | 0.10190 |
| G+C content | 0.35196 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -29.56 |
| Energy contribution | -30.16 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.89 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17538825 108 - 23011544 UACAAAUGGGUUCAAUAGGUGGGAACUGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACCAAGUCAAGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUG ..((((((((....(((((((((((((((((..((.....))..))))))....((((.(((((...))))).))))............))))))))))))))))))) ( -31.10, z-score = -3.89, R) >droEre2.scaffold_4845 15880465 102 + 22589142 ------UGGUUUGAAUAGGUGGGAACCGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGUGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUG ------......(((((((((((((((((((..((.....))..))))))....((((.(((((...))))).))))............)))))))))))))...... ( -28.00, z-score = -2.93, R) >droYak2.chr2R 17579705 108 + 21139217 UGCAAAUGGGUUCAAUAGGUGGGAACCGAAACAUAUGACUUCAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGUGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUG ..((((((((....(((((((((((((((((...(((....)))))))))....((((.(((((...))))).))))............))))))))))))))))))) ( -33.20, z-score = -4.12, R) >droSec1.super_7 1180450 108 - 3727775 UACAAAUGGGUUCAAUAGGUGGGAACCGAAAUAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCACGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUG ..((((((((....(((((((((((..........(((((((....(((((.......))))).)))))))..(((((...)))))...))))))))))))))))))) ( -30.70, z-score = -3.79, R) >droSim1.chr2L 17242876 108 - 22036055 UACAAAUGGGUUCAAUAGGUGGGAACCGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUG ..((((((((....(((((((((((((((((..((.....))..))))))....((((.(((((...))))).))))............))))))))))))))))))) ( -33.30, z-score = -4.72, R) >consensus UACAAAUGGGUUCAAUAGGUGGGAACCGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAUUUG ..(((((((....((((((((((((((((((.............))))))....((((.(((((...))))).))))............))))))))))))))))))) (-29.56 = -30.16 + 0.60)

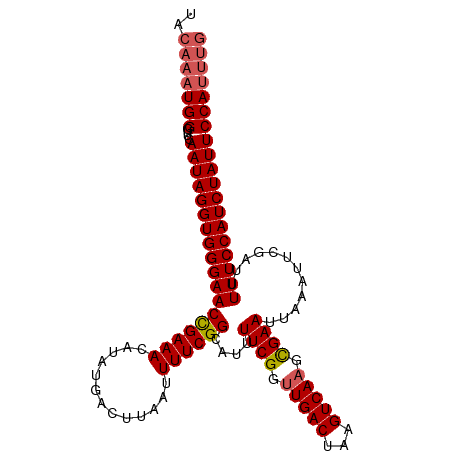
| Location | 17,538,828 – 17,538,936 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Shannon entropy | 0.12195 |
| G+C content | 0.36684 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -18.98 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17538828 108 + 23011544 AUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCUUGACUUGGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCAGUUCCCACCUAUUGAACCCAUUUGUAGCC ..((.((((((((.....(((.((((...(((.(((((...))))).)))))))..)))................((((((........)))))).))))))))..)) ( -19.60, z-score = -0.54, R) >droEre2.scaffold_4845 15880468 99 - 22589142 AUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCACUUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCGGUUCCCACCUAUUCAAACCA--------- ...((((((.(((...(((((((.......((..(((((((((....((.......))...)))))))))..)).))))))).))).))))))......--------- ( -23.60, z-score = -2.91, R) >droYak2.chr2R 17579708 108 - 21139217 AUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCACUUGACUUAGUCAACCGAAAAUGCCGAAAAUGAAGUCAUAUGUUUCGGUUCCCACCUAUUGAACCCAUUUGCAGCC (((((((((.(((...............(((..(((((...)))))..)))...((((((((((....)))...)))))))..))).)))))....))))........ ( -19.90, z-score = -0.60, R) >droSec1.super_7 1180453 108 + 3727775 AUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCGUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUAUUUCGGUUCCCACCUAUUGAACCCAUUUGUAGCC ..((.(((((((((((...(((((...))))).((((((((((....((.......))...))))))))))...))))(((((.........))))))))))))..)) ( -22.20, z-score = -1.44, R) >droSim1.chr2L 17242879 108 + 22036055 AUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCUUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCGGUUCCCACCUAUUGAACCCAUUUGUAGCC (((((((((.(((...............((((.(((((...))))).))))...(((((((..((.....))..)))))))..))).)))))....))))........ ( -21.60, z-score = -1.26, R) >consensus AUGGAAUAGAUGGAAAAAUCGAAUUUAAUUCGCUUGACUUAGUCAACCGAAAAUGCCGAAAAUUAAGUCAUAUGUUUCGGUUCCCACCUAUUGAACCCAUUUGUAGCC .((((((((.(((...(((((((.......((..(((((((((....((.......))...)))))))))..)).))))))).))).)))))....)))......... (-18.98 = -19.22 + 0.24)


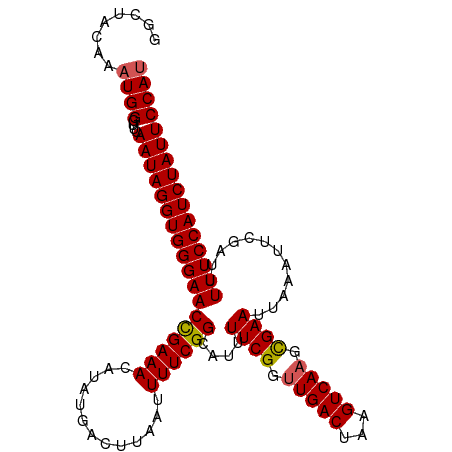
| Location | 17,538,828 – 17,538,936 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Shannon entropy | 0.12195 |
| G+C content | 0.36684 |
| Mean single sequence MFE | -27.64 |
| Consensus MFE | -26.54 |
| Energy contribution | -26.54 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17538828 108 - 23011544 GGCUACAAAUGGGUUCAAUAGGUGGGAACUGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACCAAGUCAAGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAU ........(((((....(((((((((((((((((..((.....))..))))))....((((.(((((...))))).))))............)))))))))))))))) ( -26.50, z-score = -2.05, R) >droEre2.scaffold_4845 15880468 99 + 22589142 ---------UGGUUUGAAUAGGUGGGAACCGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGUGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAU ---------......(((((((((((((((((((..((.....))..))))))....((((.(((((...))))).))))............)))))))))))))... ( -28.00, z-score = -3.06, R) >droYak2.chr2R 17579708 108 + 21139217 GGCUGCAAAUGGGUUCAAUAGGUGGGAACCGAAACAUAUGACUUCAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGUGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAU ........(((((....(((((((((((((((((...(((....)))))))))....((((.(((((...))))).))))............)))))))))))))))) ( -28.90, z-score = -2.35, R) >droSec1.super_7 1180453 108 - 3727775 GGCUACAAAUGGGUUCAAUAGGUGGGAACCGAAAUAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCACGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAU ........(((((....(((((((((((..........(((((((....(((((.......))))).)))))))..(((((...)))))...)))))))))))))))) ( -26.10, z-score = -1.93, R) >droSim1.chr2L 17242879 108 - 22036055 GGCUACAAAUGGGUUCAAUAGGUGGGAACCGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAU ........(((((....(((((((((((((((((..((.....))..))))))....((((.(((((...))))).))))............)))))))))))))))) ( -28.70, z-score = -2.87, R) >consensus GGCUACAAAUGGGUUCAAUAGGUGGGAACCGAAACAUAUGACUUAAUUUUCGGCAUUUUCGGUUGACUAAGUCAAGCGAAUUAAAUUCGAUUUUUCCAUCUAUUCCAU ........((((....((((((((((((((((((.............))))))....((((.(((((...))))).))))............)))))))))))))))) (-26.54 = -26.54 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:10 2011