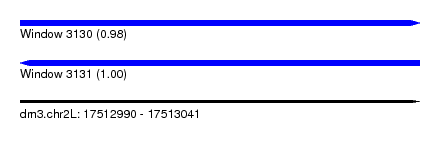
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,512,990 – 17,513,041 |
| Length | 51 |
| Max. P | 0.995181 |

| Location | 17,512,990 – 17,513,041 |
|---|---|
| Length | 51 |
| Sequences | 12 |
| Columns | 51 |
| Reading direction | forward |
| Mean pairwise identity | 80.36 |
| Shannon entropy | 0.40777 |
| G+C content | 0.56209 |
| Mean single sequence MFE | -20.18 |
| Consensus MFE | -12.87 |
| Energy contribution | -12.98 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
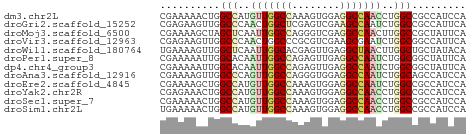
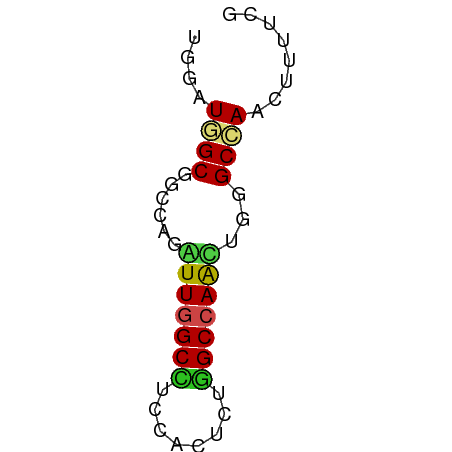
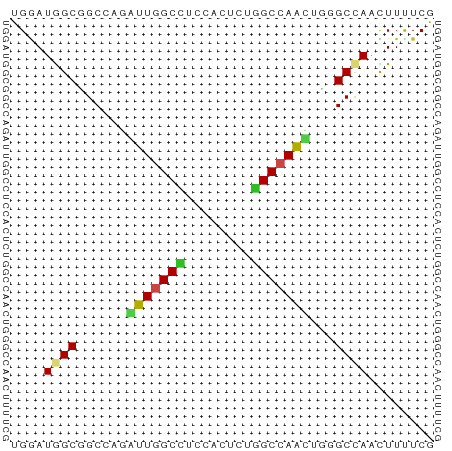
>dm3.chr2L 17512990 51 + 23011544 CGAAAAACUGGCCAUGUUGGCCAAAGUGGAGGCCAACCUGGCCGCCAUCCA .........(((((.(((((((........))))))).)))))........ ( -23.80, z-score = -3.29, R) >droGri2.scaffold_15252 13852230 51 + 17193109 CGAGAAGUUGGCCCAACUGGCUCGAGUCGAAGCCAAUCUGGCCGCCAUUCA ...(((..(((((....(((((((...)).)))))....)))))...))). ( -17.20, z-score = -1.45, R) >droMoj3.scaffold_6500 21359677 51 + 32352404 CGAAAAGCUAGCUCAAUUGGCCAGGGUCGAGGCCAACUUGGCCGCUAUUCA .(((.(((..((.(((((((((........)))))).))))).))).))). ( -18.70, z-score = -1.92, R) >droVir3.scaffold_12963 16429435 51 + 20206255 CGAGAAGUUGGCCCAACUGGCCCGCGUCGAAGCGAAUCUGGCGGCCAUUCA ...(((..(((((...(..(..(((......)))...)..).)))))))). ( -15.30, z-score = 0.14, R) >droWil1.scaffold_180764 2945181 51 - 3949147 UGAAAAGUUGGCUCAAUUGGCACGAGUUGAGGCUAACUUGGCUGCUAUACA .(..((((((((((((((......)))))).))))))))..)......... ( -15.10, z-score = -1.97, R) >droPer1.super_8 1831227 51 - 3966273 CGAAAAAUUGGCACAAUUGGCCAGAGUUGAGGCCAAUCUGGCGGCUAUUCA .(((....((((.(((((((((........))))))).))...))))))). ( -15.90, z-score = -1.71, R) >dp4.chr4_group3 768199 51 - 11692001 CGAAAAAUUGGCACAAUUGGCCAGAGUUGAGGCCAAUCUGGCGGCUAUUCA .(((....((((.(((((((((........))))))).))...))))))). ( -15.90, z-score = -1.71, R) >droAna3.scaffold_12916 12812903 51 - 16180835 CGAAAAGUUGGCCCAGUUGGCCAGGGUGGAGGCCAAUCUGGCAGCCAUCCA ........(((((.....)))))((((((..(((.....)))..)))))). ( -24.90, z-score = -2.87, R) >droEre2.scaffold_4845 15853772 51 - 22589142 CGAAAAGCUGGCCAUGUUGGCCAAAGUGGAGGCCAAUCUGGCCGCCAUCCA ......((.(((((.(((((((........))))))).)))))))...... ( -23.70, z-score = -2.93, R) >droYak2.chr2R 17552784 51 - 21139217 CGAGAAACUGGCCAUGUUGGCCAAAGUGGAGGCCAACCUGGCCGCCAUCCA .(.((...((((((.(((((((........))))))).))))))...))). ( -24.00, z-score = -3.17, R) >droSec1.super_7 1154024 51 + 3727775 CGAAAAACUGGCCAUGUUGGCCAAAGUGGAGGCCAACCUGGCCGCCAUCCA .........(((((.(((((((........))))))).)))))........ ( -23.80, z-score = -3.29, R) >droSim1.chr2L 17217231 51 + 22036055 UGAAAAACUGGCCAUGUUGGCCAAAGUGGAGGCCAACCUGGCCGCCAUCCA .........(((((.(((((((........))))))).)))))........ ( -23.80, z-score = -3.28, R) >consensus CGAAAAACUGGCCCAAUUGGCCAGAGUGGAGGCCAACCUGGCCGCCAUCCA ..........(((..(((((((........)))))))..)))......... (-12.87 = -12.98 + 0.11)

| Location | 17,512,990 – 17,513,041 |
|---|---|
| Length | 51 |
| Sequences | 12 |
| Columns | 51 |
| Reading direction | reverse |
| Mean pairwise identity | 80.36 |
| Shannon entropy | 0.40777 |
| G+C content | 0.56209 |
| Mean single sequence MFE | -20.92 |
| Consensus MFE | -13.00 |
| Energy contribution | -12.89 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.995181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
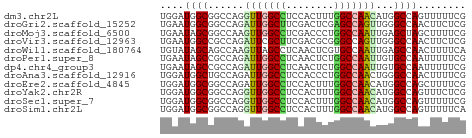
>dm3.chr2L 17512990 51 - 23011544 UGGAUGGCGGCCAGGUUGGCCUCCACUUUGGCCAACAUGGCCAGUUUUUCG .(((..(((((((.(((((((........))))))).))))).))..))). ( -27.50, z-score = -4.25, R) >droGri2.scaffold_15252 13852230 51 - 17193109 UGAAUGGCGGCCAGAUUGGCUUCGACUCGAGCCAGUUGGGCCAACUUCUCG .(((.(..((((.(((((((((......))))))))).))))..))))... ( -21.80, z-score = -2.80, R) >droMoj3.scaffold_6500 21359677 51 - 32352404 UGAAUAGCGGCCAAGUUGGCCUCGACCCUGGCCAAUUGAGCUAGCUUUUCG .(((.((((((..((((((((........))))))))..))).))).))). ( -22.60, z-score = -3.97, R) >droVir3.scaffold_12963 16429435 51 - 20206255 UGAAUGGCCGCCAGAUUCGCUUCGACGCGGGCCAGUUGGGCCAACUUCUCG .((((((((((..(.(((((......))))))..))..)))))..)))... ( -13.40, z-score = 0.68, R) >droWil1.scaffold_180764 2945181 51 + 3949147 UGUAUAGCAGCCAAGUUAGCCUCAACUCGUGCCAAUUGAGCCAACUUUUCA .........(..(((((.((.((((..........)))))).)))))..). ( -5.70, z-score = 0.61, R) >droPer1.super_8 1831227 51 + 3966273 UGAAUAGCCGCCAGAUUGGCCUCAACUCUGGCCAAUUGUGCCAAUUUUUCG .(((.....((..((((((((........))))))))..))......))). ( -15.10, z-score = -2.48, R) >dp4.chr4_group3 768199 51 + 11692001 UGAAUAGCCGCCAGAUUGGCCUCAACUCUGGCCAAUUGUGCCAAUUUUUCG .(((.....((..((((((((........))))))))..))......))). ( -15.10, z-score = -2.48, R) >droAna3.scaffold_12916 12812903 51 + 16180835 UGGAUGGCUGCCAGAUUGGCCUCCACCCUGGCCAACUGGGCCAACUUUUCG .((.((((..((((.((((((........)))))))))))))).))..... ( -22.10, z-score = -2.92, R) >droEre2.scaffold_4845 15853772 51 + 22589142 UGGAUGGCGGCCAGAUUGGCCUCCACUUUGGCCAACAUGGCCAGCUUUUCG .(((.((((((((..((((((........))))))..))))).))).))). ( -27.10, z-score = -4.51, R) >droYak2.chr2R 17552784 51 + 21139217 UGGAUGGCGGCCAGGUUGGCCUCCACUUUGGCCAACAUGGCCAGUUUCUCG .((..((((((((.(((((((........))))))).))))).)))..)). ( -25.60, z-score = -3.40, R) >droSec1.super_7 1154024 51 - 3727775 UGGAUGGCGGCCAGGUUGGCCUCCACUUUGGCCAACAUGGCCAGUUUUUCG .(((..(((((((.(((((((........))))))).))))).))..))). ( -27.50, z-score = -4.25, R) >droSim1.chr2L 17217231 51 - 22036055 UGGAUGGCGGCCAGGUUGGCCUCCACUUUGGCCAACAUGGCCAGUUUUUCA .(((..(((((((.(((((((........))))))).))))).))..))). ( -27.50, z-score = -3.97, R) >consensus UGGAUGGCGGCCAGAUUGGCCUCCACUCUGGCCAACUGGGCCAACUUUUCG ....((((......(((((((........)))))))...))))........ (-13.00 = -12.89 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:07 2011