| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,512,546 – 17,512,641 |
| Length | 95 |
| Max. P | 0.736465 |

| Location | 17,512,546 – 17,512,641 |
|---|---|
| Length | 95 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.26 |
| Shannon entropy | 0.44649 |
| G+C content | 0.49357 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -15.38 |
| Energy contribution | -14.57 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.736465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
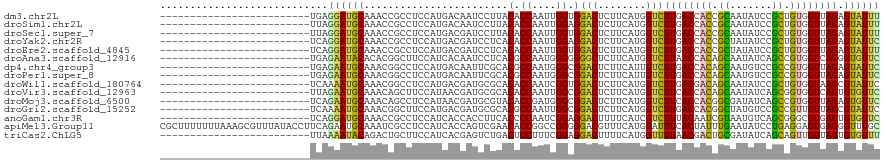
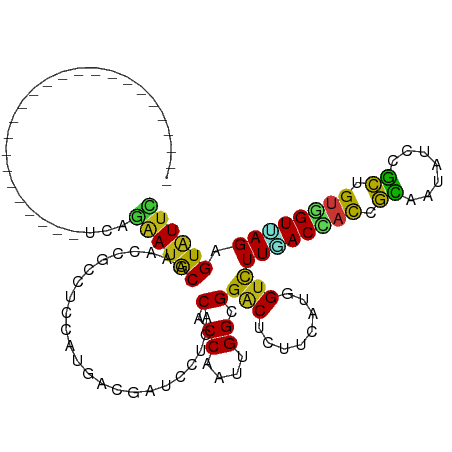
>dm3.chr2L 17512546 95 - 23011544 -------------------------UGAGGAUGCAAACCGCCUCCAUGACAAUCCUUACACCAAUUGGUGGACUCUUCAUGGUCUUGACCACCGCAAUAUCCGCUGUGGUUAGAGUAUUU -------------------------...((((((.........((((((.........((((....))))......))))))(((.((((((.((.......)).))))))))))))))) ( -26.86, z-score = -1.68, R) >droSim1.chr2L 17216787 95 - 22036055 -------------------------UUAGGAUGCAAACCGCCUCCAUGACAAUCCUUACACCAAUUGGAGGACUCUUCAUGGUCUUGACCACCGCAAUAUCCGCUGUGGUUAGAGUAUUU -------------------------...((((((.........((((((...(((((.((.....)))))))....))))))(((.((((((.((.......)).))))))))))))))) ( -26.50, z-score = -2.01, R) >droSec1.super_7 1153580 95 - 3727775 -------------------------UUAGGAUGCAAACCGCCUCCAUGACGAUCCUUACACCAAUUGGUGGACUCUUCAUGGUCUUGACCACCGCAAUAUCCGCUGUGGUUAGAGUAUUU -------------------------...((((((.........((((((.(((((....(((....)))))).)).))))))(((.((((((.((.......)).))))))))))))))) ( -29.00, z-score = -2.54, R) >droYak2.chr2R 17552340 95 + 21139217 -------------------------UCAGGAUGCAAACCGCCUCCAUGACGAUCCUCACACCAAUUGGAGGACUCUUCAUGGUCUUGACCACCGCUAUAUCCGCUGUGGUUAGAGUAUUC -------------------------...((((((.........((((((.(((((((.((.....))))))).)).))))))(((.((((((.((.......)).))))))))))))))) ( -33.40, z-score = -3.49, R) >droEre2.scaffold_4845 15853328 95 + 22589142 -------------------------UCAGGAUGCAAACCGCCUCCAUGACGAUCCUCACACCAAUUGGUGGACUCUUCAUGGUCUUGACCACCGCUAUAUCCGCUGUGGUUAGAGUAUUU -------------------------...((((((.........((((((.((...((.((((....)))))).)).))))))(((.((((((.((.......)).))))))))))))))) ( -29.90, z-score = -2.46, R) >droAna3.scaffold_12916 12812459 95 + 16180835 -------------------------UGAGAAUACACACGGCUUCCAUCACAAUCCUCACGCCAAUGGGGGGGCUCUUCAUGGUCUUAACCACAGCAAUAUCAGCCGUGGUCAGGGUGUUC -------------------------...((((((.(((((((..........((((((......)))))).(((.....((((....)))).)))......)))))))......)))))) ( -26.20, z-score = -0.64, R) >dp4.chr4_group3 767755 95 + 11692001 -------------------------UGAGAAUGCAAACGGCCUCCAUGACAAUUCGCACGCCAAUGGGCGGACUCUUCAUUGUCUUGACCACAGCAAUGUCCGCCGUGGUUAGAGUAUUC -------------------------...((((((.............((((((..(..((((....))))..).....))))))((((((((.((.......)).)))))))).)))))) ( -28.30, z-score = -1.10, R) >droPer1.super_8 1830783 95 + 3966273 -------------------------UGAGAAUGCAAACGGCCUCCAUGACAAUUCGCACGCCAAUGGGCGGACUCUUCAUUGUCUUGACCACAGCAAUGUCCGCCGUGGUUAGAGUAUUC -------------------------...((((((.............((((((..(..((((....))))..).....))))))((((((((.((.......)).)))))))).)))))) ( -28.30, z-score = -1.10, R) >droWil1.scaffold_180764 2944737 95 + 3949147 -------------------------UCAAAAUGCAAACGGCCUCCAUGACGAUGCGCACACCAAUCGGUGGACUCUUCAUGGUCUUGACGACAGCAAUAUCCGCUGUGGUAAGCGUAUUC -------------------------.....((((...(((...((((((.((...(..((((....))))..))).))))))..)))((.(((((.......))))).))..)))).... ( -27.30, z-score = -1.24, R) >droVir3.scaffold_12963 16428991 95 - 20206255 -------------------------UUAGAAUGCAAACAGCUUCCAUAACGAUGCGCACACCAAUUGGCGGACUCUUCAUGGUCUUCACCACAGCAAUAUCAGCGGUGGUCAGUGUGUUC -------------------------...(((.((.....))))).........((((((........(.(((((......))))).)(((((.((.......)).)))))..)))))).. ( -22.00, z-score = 0.10, R) >droMoj3.scaffold_6500 21359233 95 - 32352404 -------------------------UCAGAAUGCAAACAGCCUCCAUAACGAUGCGUACACCGAUGGGCGGACUCUUCAUGGUCUUCACCACGGCGAUAUCAGCCGUGGUUAGAGUGUUC -------------------------...((((((....(((((((((..((.((....)).))))))).)).))........(((..((((((((.......)))))))).))))))))) ( -28.20, z-score = -0.85, R) >droGri2.scaffold_15252 13851786 95 - 17193109 -------------------------UCAAAAUGCAAACGGCUUCCAUGACGAUGCGCACGCCAAUUGGCGGACUCUUCAUGGUCUUGACCACGGCUAUGUCCGCCGUUGUUAGCGUAUUC -------------------------.....((((.((((((..((((((.((...(..((((....))))..))).))))))....).))(((((.......))))).))).)))).... ( -30.50, z-score = -1.54, R) >anoGam1.chr3R 37046128 95 - 53272125 -------------------------UCAGGAUGCAAACCGCCUCCAUCACCACCUUCACCCCAAUCGGAGGACUUUUCAUCGUCUUUACAAUCGUAAUGUCAGCGGGCGUGAGUGUGUUC -------------------------...(((.((.....)).))).....(((.((((((((.....((((((........))))))......((.......))))).))))).)))... ( -24.80, z-score = -1.04, R) >apiMel3.Group11 4433329 120 - 12576330 CGCUUUUUUUAAAGCGUUUAUACCUUCAGAAUGCAAAUCGCCUCCAUCACCAGUCGAACACCGGCCGGGGGACGUUUCAUGGAUUUCACUAUUUGAAUAUCCUGAGGAGUGAGGGUUUGC ((((((....)))))).....(((((((....((.....))((((.((.((.((((.....)))).))..))....(((.((((((((.....)))).)))))))))))))))))).... ( -34.70, z-score = -1.50, R) >triCas2.ChLG5 16918580 95 - 18847211 -------------------------UUAAAAUACAGACUGCUUCCAUCACGAGUCUGACUCCUUUCGGAGGACUUUUCAUGGUUUUAACGACUGCGAUAUCAGCAGUUGUUAGUGUGUUU -------------------------...(((((((........((((.(.(((((...((((....))))))))).).))))..(((((((((((.......)))))))))))))))))) ( -30.50, z-score = -3.43, R) >consensus _________________________UCAGAAUGCAAACCGCCUCCAUGACGAUCCUCACACCAAUUGGCGGACUCUUCAUGGUCUUGACCACCGCAAUAUCCGCUGUGGUUAGAGUAUUC ............................((((((........................(.((....)).)(((........)))((((((((.((.......)).)))))))).)))))) (-15.38 = -14.57 + -0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:05 2011