| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,464,152 – 17,464,262 |
| Length | 110 |
| Max. P | 0.856048 |

| Location | 17,464,152 – 17,464,262 |
|---|---|
| Length | 110 |
| Sequences | 13 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 70.98 |
| Shannon entropy | 0.62380 |
| G+C content | 0.47926 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.59 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
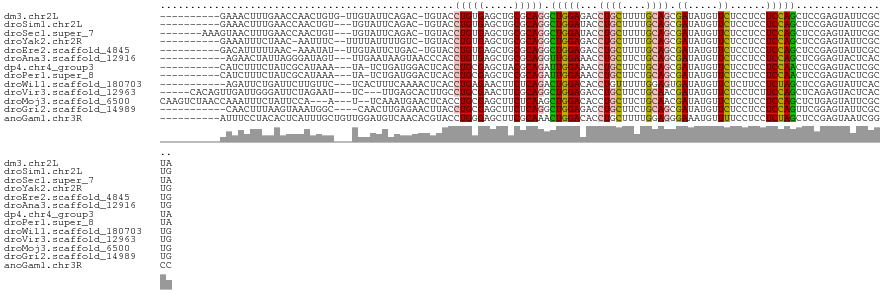
>dm3.chr2L 17464152 110 + 23011544 ----------GAAACUUUGAACCAACUGUG-UUGUAUUCAGAC-UGUACCUGUGAGCUGCGCAGGCUGGAGACCUGCUUUUGCAGCGAUAUGUUCUCCUCCUCCAGCUCCGAGUAUUCGCUA ----------(((((((.((.....(((((-(.((...(((..-.....)))...)).))))))(((((((..((((....)))).((.....)).....))))))))).)))).))).... ( -33.90, z-score = -2.12, R) >droSim1.chr2L 17166417 108 + 22036055 ----------GAAACUUUGAACCAACUGU---UGUAUUCAGAC-UGUACCUGUGAGCUGCGCAGGCUGGAUACCUGCUUUUGCAGCGAUAUGUUCUCCUCCUCCAGCUCCGAGUAUUCGCUG ----------...................---......(((..-.((((.((.((((((.(.(((..(((...((((....)))).((.....))))).)))))))))))).))))...))) ( -27.60, z-score = -0.84, R) >droSec1.super_7 1105458 111 + 3727775 -------AAAGUAACUUUGAACCAACUGU---UGUAUUCAGAC-UGUACCUGUGAGCUGCGCAGGCUGGAUACCUGCUUUUGCAGCGAUAUGUUCUCCUCCUCCAGCUCCGAGUAUUCGCUA -------..(((...((((((...((...---.)).)))))).-.((((.((.((((((.(.(((..(((...((((....)))).((.....))))).)))))))))))).))))..))). ( -26.90, z-score = -0.35, R) >droYak2.chr2R 17503607 108 - 21139217 ----------GAAAUUUCUAAC-AAUUUC--UUUUAUUUUGUC-UGUACCUGUGAGCUGCGCAGGCUGGAGACCUGCUUUUGCAGCGAUAUGUUCUCCUCCUCCAGCUCCGAGUAUUCGCUG ----------((((((......-))))))--.........((.-.((((.((.((((((.(.(((..((((((((((....)))).)......))))).)))))))))))).))))..)).. ( -31.50, z-score = -2.87, R) >droEre2.scaffold_4845 15803427 108 - 22589142 ----------GACAUUUUUAAC-AAAUAU--UUGUAUUCUGAC-UGUACCUGUGAGCUGCGCAGGCUGGAGACCUGCUUUUGCAGCGAUAUGUUCUCCUCCUCCAGCUCCGAGUAUUCGCUG ----------..........((-((....--)))).....((.-.((((.((.((((((.(.(((..((((((((((....)))).)......))))).)))))))))))).)))))).... ( -28.30, z-score = -1.42, R) >droAna3.scaffold_12916 12764210 108 - 16180835 -----------AGAACUAUUAGGGAUAGU---UUGAAUAAGUAACCCACCUGUGAGCUGCGCAGGUUGGAAACCUGCUUCUGCAGCGAUAUGUUCUCCUCCUCCAGCUCGGAGUACUCACUG -----------(((((((((.(((.((.(---(.....)).)).)))........(((((((((((.....))))))....))))))))).)))))....((((.....))))......... ( -32.00, z-score = -1.21, R) >dp4.chr4_group3 11008521 108 + 11692001 ----------CAUCUUUCUAUCGCAUAAA---UA-UCUGAUGGACUCACCUGCGAGCUACGCAGAUUGGAAACCUGCUUCUGCAGCGAUAUGUUCUCCUCCUCCAACUCCGAGUACUCGCUA ----------......(((((((.((...---.)-).))))))).......(((((.(((.(.(((((((...((((....)))).((.....))......))))).)).).)))))))).. ( -26.30, z-score = -1.98, R) >droPer1.super_8 1749353 108 - 3966273 ----------CAUCUUUCUAUCGCAUAAA---UA-UCUGAUGGACUCACCUGCGAGCUCCGCAGAUUGGAAACCUGCUUCUGCAGCGAUAUGUUCUCCUCCUCCAACUCCGAGUACUCGCUA ----------......(((((((.((...---.)-).))))))).......(((((..(((.((.(((((...((((....)))).((.....))......))))))).)).)..))))).. ( -26.10, z-score = -1.83, R) >droWil1.scaffold_180703 1576592 108 - 3946847 -----------AGAUUCUGAUUCUUGUUC---UCACUUUCAAAACUCACCUGAGAACUUCUCAGACUGGACACCUGUUUUUGGAGUGAUAUGUUCUCUUCCUCUAGCUCCGAGUAUUCACUG -----------....(((((.....((((---(((...............)))))))...))))).((((.((.((...((((((.((..........))))))))...)).)).))))... ( -19.06, z-score = 0.99, R) >droVir3.scaffold_12963 10617038 111 - 20206255 -----CACAGUUGAUUGGGAUUCUAGAAU---UC---UUGAGCACUUGCCUGCGAACUUCGCAGGCUGGAGACCUGCUUCUGCAACGAUAUGUUCUCCUCUUCCAGCUCAGAGUACUCACUG -----..((((.((((((....)))).((---((---(.((((....(((((((.....))))))).(((((.((((....))).......).))))).......)))))))))..)))))) ( -35.11, z-score = -1.81, R) >droMoj3.scaffold_6500 1677629 114 + 32352404 CAAGUCUAACCAAAUUUCUAUUCCA---A---U--UCAAAUGAACUCACCUGCGAGCUUCUCAAGCUGGACACCUGCUUCUGCAACGAUAUGUUCUCCUCCUCCAGCUCUGAGUAUUCGCUG .........................---.---.--................(((((...(((((((((((....(((....)))..((.....))......))))))).))))..))))).. ( -24.00, z-score = -2.46, R) >droGri2.scaffold_14989 497 107 + 50547 -----------CAACUUUAAGUAAAUGGC----CAACUUGAGAACUUACCUGCGAGCUUCUCAGGCUGGAGACCUGCUUCUGCAGCGAUAUGUUCUCCUCCUCCAGUUCGGAGUAUUCGCUG -----------........((..((((.(----(..(((((((((((......))).))))))))((((((..((((....)))).((.....)).....))))))...))..))))..)). ( -30.10, z-score = -1.04, R) >anoGam1.chr3R 51393689 112 + 53272125 ----------AUUUCCUACACUCAUUUGCUGUUGGAUGUCAACACGUACCUGGGAGCUUCGCAAACUGGACACCUGCUUUUGGAGGGAAAUGUUUUCCUCCUCUAGCUCCGAGUAAUCGGCC ----------...(((....((((..((((((((.....))))).)))..)))).((...)).....))).....(((...((((((((....))))))))...))).((((....)))).. ( -32.40, z-score = -1.41, R) >consensus __________GAAAUUUUUAACCAAUUGU___UGUAUUUAGACACGUACCUGUGAGCUGCGCAGGCUGGAGACCUGCUUUUGCAGCGAUAUGUUCUCCUCCUCCAGCUCCGAGUAUUCGCUG .................................................(((((.....))))).(((((...((((....)))).((.....))......)))))................ (-12.69 = -12.59 + -0.10)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:48:01 2011